Hi! My name is Michal and I'm currently studying Mathematics and Computer Science at the University of Oxford. I have just finished the second year of my degree and joined Genestack for a summer internship.
Hi! My name is Michal and I'm currently studying Mathematics and Computer Science at the University of Oxford. I have just finished the second year of my degree and joined Genestack for a summer internship.
I'm currently developing a Genestack application which will load and process the metadata of over 40,000 public experiments from various databases in order to make them available on the platform. When the data will be released, all of these experiments will be directly integrated into Genestack. You'll be able to access them and search through them from the File Manager, and include them seamlessly into your data flows - the same way you would work with your own data.
This is a continuation of a project that has been running at Genestack for some time. The goal is to make all the large scale genLife Science Datasets available for analysis in one place, seamlessly. We have been indexing all the sequencing data from four major repositories: SRA, ENA, ArrayExpress and GEO. Every indexed experiment and assay from these databases can be found on Genestack and can be used in analyses right away.
These databases altogether contain a very large number of experiments with over a million assays, which presents a couple of challenges. First of all as it is very difficult to keep this much data well organised, so there are a few discrepancies, such as missing files, that I have to deal with. Furthermore, a lot of experiments are contained in more than one database, so we need to find the best way to combine metadata from all of them to give the users as much information as possible.
We started with the SRA database at NCBI and I recently added parsers for ENA and ArrayExpress at the EBI. I'm now testing and debugging them.
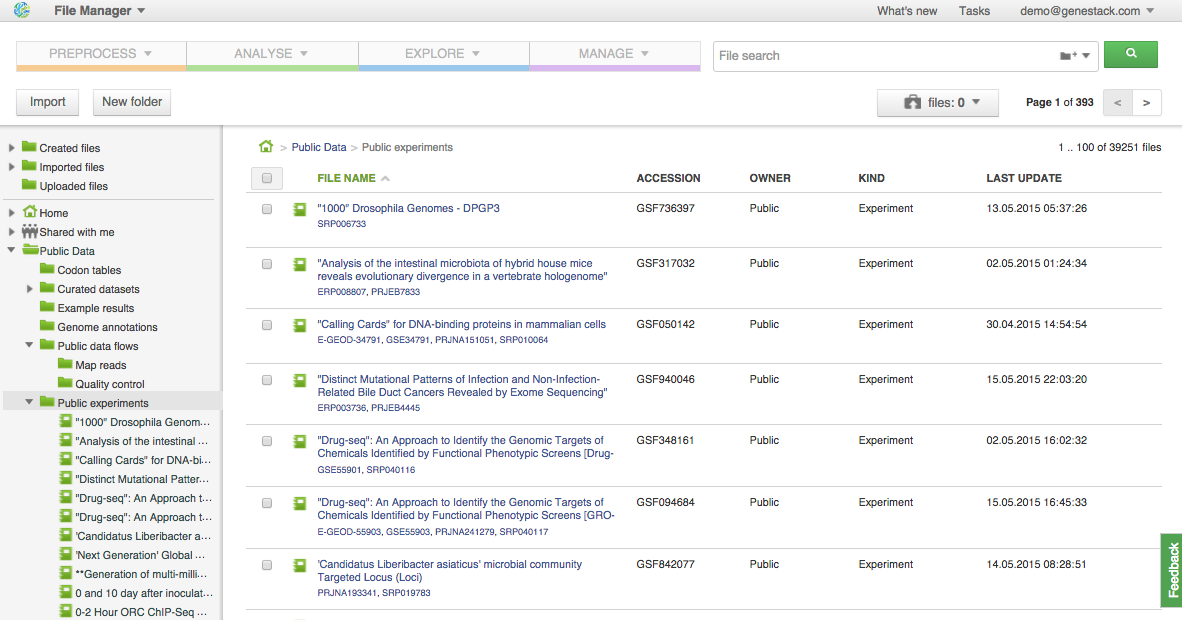
At the moment, there are various public experiments from SRA, BioProject, ArrayExpress and other databases already imported, including datasets from such large-scale projects as the 1000 genomes, HapMap and ENCODE:
To sum up, the primary aim of the project is to have all of the public sequencing experiments from the databases mentioned above imported on the platform by the end of the summer. Furthermore, we want to get as much up to date metadata about the experiments and their assays as possible and organise it in a way to make it easy for the users to find the experiment they are looking for.
Resources: