 Genestack's booth at PAG 2017 As always, we're amazed by the complexities of plant and animal genetics. But we're equally encouraged by the amazing progress experienced in this field. One such effort is improved whole genome sequencing using better technologies and strategies: during PAG, we learned about how the first public genome sequence for the most widely cultivated coffee species, Coffea arabica, was de novo assembled using high coverage PACBio long reads, error-corrected using Illumina reads, and was further scaffolded and improved using optical maps. Progress has also been made in constructing pangenomes for new species, including Brassica Napus Canola and Oleracea, wheat, and maize, to help capture the variability of genes between different varieties of a species. The availability of pangenomes facilitates a comprehensive discovery of genes and variants related to key agrigenomics traits. Genome editing is gaining wider adoption, and many talks were showcasing the different methods and their accomplishments: from successfully using CRISPR/Cas9 in developing herbicide tolerant cassava, to evidence that suggests TALENs may be a better approach for wheat genome editing. As these technologies mature, high-throughput phenotyping and genomic selection methods become increasingly more relevant and applicable. Other technologies that captured our interests are linked-reads and single cell gene expression developments by 10x. Linked-reads have the benefits of accuracy and detection power of short reads, but also include long-range information. This makes them useful in detecting large structural variations, including translocation and more complex events involving multiple break points. Furthermore, single-cell gene expression profiling has now become more scalable than ever -- 10x have released a new dataset that covers a million mouse brain cells. We are looking forward to analysing this dataset through our single-cell transcriptomic pipeline! Our workshop, "Applying High-throughput Bioinformatics Technologies to Crop Research", was held on Thursday, 17th January. We were pleased to meet with a diverse attendee group composed of researchers from academia and industry. Keywan started the workshop by providing an Overview of KnetMiner, expanding his earlier talk and showcasing the key features and biological insights using KnetMiner through a live demo. He demonstrated how KnetMiner can rapidly search and evaluate a vast amount of heterogeneous relations and knowledge types to determine if direct or indirect links between genes and trait-related keywords can be established, and how users can explore the most relevant genes through interactive genome and network maps.
Genestack's booth at PAG 2017 As always, we're amazed by the complexities of plant and animal genetics. But we're equally encouraged by the amazing progress experienced in this field. One such effort is improved whole genome sequencing using better technologies and strategies: during PAG, we learned about how the first public genome sequence for the most widely cultivated coffee species, Coffea arabica, was de novo assembled using high coverage PACBio long reads, error-corrected using Illumina reads, and was further scaffolded and improved using optical maps. Progress has also been made in constructing pangenomes for new species, including Brassica Napus Canola and Oleracea, wheat, and maize, to help capture the variability of genes between different varieties of a species. The availability of pangenomes facilitates a comprehensive discovery of genes and variants related to key agrigenomics traits. Genome editing is gaining wider adoption, and many talks were showcasing the different methods and their accomplishments: from successfully using CRISPR/Cas9 in developing herbicide tolerant cassava, to evidence that suggests TALENs may be a better approach for wheat genome editing. As these technologies mature, high-throughput phenotyping and genomic selection methods become increasingly more relevant and applicable. Other technologies that captured our interests are linked-reads and single cell gene expression developments by 10x. Linked-reads have the benefits of accuracy and detection power of short reads, but also include long-range information. This makes them useful in detecting large structural variations, including translocation and more complex events involving multiple break points. Furthermore, single-cell gene expression profiling has now become more scalable than ever -- 10x have released a new dataset that covers a million mouse brain cells. We are looking forward to analysing this dataset through our single-cell transcriptomic pipeline! Our workshop, "Applying High-throughput Bioinformatics Technologies to Crop Research", was held on Thursday, 17th January. We were pleased to meet with a diverse attendee group composed of researchers from academia and industry. Keywan started the workshop by providing an Overview of KnetMiner, expanding his earlier talk and showcasing the key features and biological insights using KnetMiner through a live demo. He demonstrated how KnetMiner can rapidly search and evaluate a vast amount of heterogeneous relations and knowledge types to determine if direct or indirect links between genes and trait-related keywords can be established, and how users can explore the most relevant genes through interactive genome and network maps.  Keywan gave an overview of KnetMiner Philipp Bayer, from the University of Western Australia, then shared his experience of using KnetMiner as an advanced analytical and decision-making tool to mine genes that lead to breeding successes in complex crop plants: Brassica oleracea, Brassica napus and Cicer arietinum. He also talked about the challenges of building these networks for the first time, where he spent a week with Keywan due to travel mishaps -- this is one objective we're hoping to achieve in Genestack: to let users build their high-quality and relevant knowledge networks without technical or domain expertise barriers. It was very useful to hear Philipp's ideas on the future of the project: ways to improve KnetMiner (e.g. machine learning approach) and how KnetMiner is an especially ideal approach to finding candidate genes in the genome editing era. The slides and audio recording of Philipp's talk are available here.
Keywan gave an overview of KnetMiner Philipp Bayer, from the University of Western Australia, then shared his experience of using KnetMiner as an advanced analytical and decision-making tool to mine genes that lead to breeding successes in complex crop plants: Brassica oleracea, Brassica napus and Cicer arietinum. He also talked about the challenges of building these networks for the first time, where he spent a week with Keywan due to travel mishaps -- this is one objective we're hoping to achieve in Genestack: to let users build their high-quality and relevant knowledge networks without technical or domain expertise barriers. It was very useful to hear Philipp's ideas on the future of the project: ways to improve KnetMiner (e.g. machine learning approach) and how KnetMiner is an especially ideal approach to finding candidate genes in the genome editing era. The slides and audio recording of Philipp's talk are available here.  Philipp shared his experience in finding candidate genes for breeding success traits in Brassicas and Cicer arietinum The last workshop session was about building and exploring knowledge networks in Genestack. Keywan and Philipp have shown how useful and powerful KnetMiner is as a discovery and interpretation tool. But, this relies on having a high-quality knowledge network. You can get useful and relevant results only if your network contains good and relevant data and relationships. At the moment, networks have been built only for nine plant species: they're updated irregularly, using only public data sources, and are hosted on servers with limited computational complexity. But, what happens if you want to study a different organism“ Or if you want to use different data sources“ This turns out to be a complicated process.
Philipp shared his experience in finding candidate genes for breeding success traits in Brassicas and Cicer arietinum The last workshop session was about building and exploring knowledge networks in Genestack. Keywan and Philipp have shown how useful and powerful KnetMiner is as a discovery and interpretation tool. But, this relies on having a high-quality knowledge network. You can get useful and relevant results only if your network contains good and relevant data and relationships. At the moment, networks have been built only for nine plant species: they're updated irregularly, using only public data sources, and are hosted on servers with limited computational complexity. But, what happens if you want to study a different organism“ Or if you want to use different data sources“ This turns out to be a complicated process. 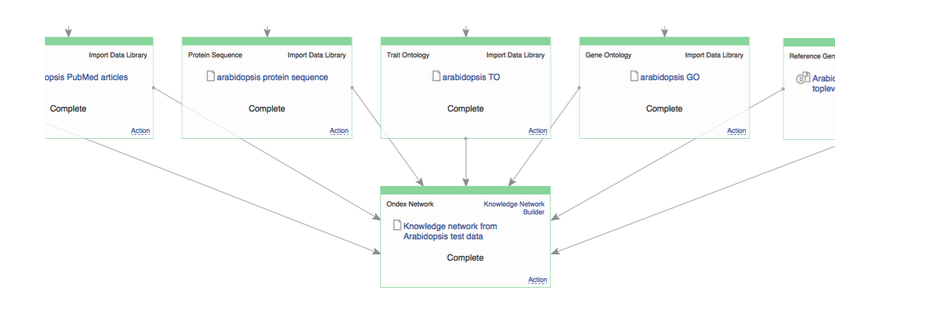 Building knowledge networks becomes straightforward in Genestack
Building knowledge networks becomes straightforward in Genestack 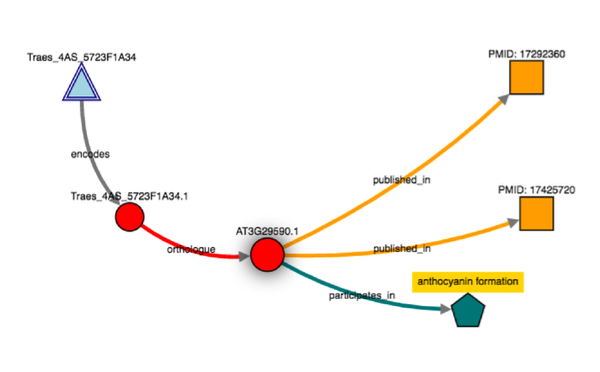 Exploring knowledge networks becomes straightforward in Genestack You start with having to gather the right Life Science Datasets: these datasets are big, scattered everywhere, and come in different flavours for different organisms. Then, you have to process and link these datasets together to form a well-defined network. But there is a steep learning curve to use Ondex, and you'd usually need a domain expert to help you. Once you have your network, there are further technical and infrastructure barriers of having to build and set-up KnetMiner server manually. KnetMiner requires high memory to hold the entire graph in memory for responsive search and interactive network visualisation. This is the documentation about the numerous technical steps you have to take to build and use KnetMiner. During our collaboration with Rothamsted research, we integrated Ondex and KnetMiner into our platform, with a purpose to streamline the entire process; facilitating end-users without any programming, bioinformatics expertise, or infrastructure resources. During the workshop, we run a Genestack live demo on:
Exploring knowledge networks becomes straightforward in Genestack You start with having to gather the right Life Science Datasets: these datasets are big, scattered everywhere, and come in different flavours for different organisms. Then, you have to process and link these datasets together to form a well-defined network. But there is a steep learning curve to use Ondex, and you'd usually need a domain expert to help you. Once you have your network, there are further technical and infrastructure barriers of having to build and set-up KnetMiner server manually. KnetMiner requires high memory to hold the entire graph in memory for responsive search and interactive network visualisation. This is the documentation about the numerous technical steps you have to take to build and use KnetMiner. During our collaboration with Rothamsted research, we integrated Ondex and KnetMiner into our platform, with a purpose to streamline the entire process; facilitating end-users without any programming, bioinformatics expertise, or infrastructure resources. During the workshop, we run a Genestack live demo on: - How to import, annotate, and combine private with public datasets. On Genestack, there are mechanisms to help users manage their data: from sharing, faceted and free-text search, to curation using controlled vocabularies and ontology terms.
- How Life Science Datasets can be built into a network easily using our simple and intuitive application
- How the generated network can be immediately explored using Genestack's KnetMiner application
- How other Genestack applications and pipelines (such as homology inference and differential expression analysis) help to make the network building and exploration process more efficient and effective
- And how the findings can always be reproduced and shared
Soon, we will release a video tutorial on how you can build and explore knowledge network in Genestack. Stay tuned for more news on the project!