Hello everyone, It's time for our next release! During the last couple of months our team has been busy developing cool new apps. Here are some of them:
Kallisto Report app
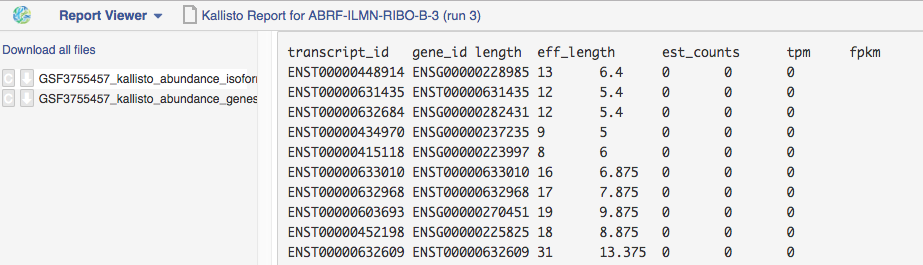
The Kallisto Report app quantifies the abundance of transcripts from RNA-Seq data without the need for alignment of individual bases. The app is based on
the Kallisto program that determines not a position of each read on the reference, but a set of potential transcripts a read could have come from, so called "pseudoalignments". With this, the app is able to analyse big RNA-seq data simply and very quickly.
Spliced mapping with STAR app
The Spliced Mapping with STAR app is our new mapper for RNA-seq data, to develop which we integrated a very popular open source software
STAR. This is an addition to our Spliced Mapping app based on TopHat tool. In comparison to TopHat, STAR works fast, at the same time being very accurate and precise. Moreover, in contrast to all our other mappers, it maps reads onto the reference transcriptome, not the genome. Another advantage of the Spliced Mapping with STAR app is that it can be used to analyse both: short and long reads, making it compatible with various sequencing platforms. Finally, the app supports two-pass alignment strategy when it runs the second alignment pass to align reads across the found splice junctions, which improves quantification of the novel splice junctions. All in all, the Spliced Mapping with STAR app can be a very good alternative to other RNA-seq aligners.
RSEM Report app
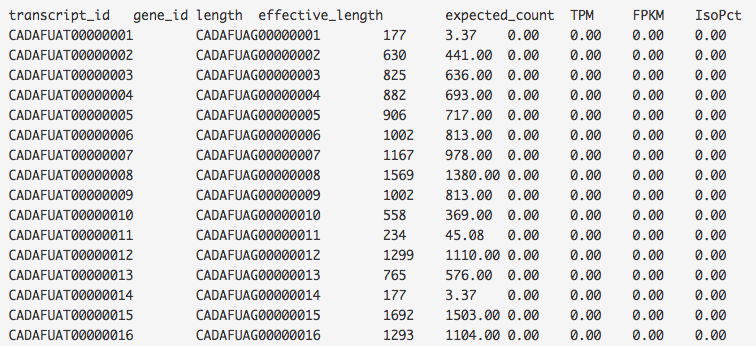
The RSEM Report app applies Expectation-Maximization algorithm to estimate gene and isoform expression levels from RNA-Seq data. Unlike the Test Differential Gene Expression app, the RSEM Report doesn't require a reference genome. Instead it uses the STAR mapper to align reads against a set of reference transcript sequences or de novo transcriptome assemblies. As a result, it generates reports outlining expression levels for genes and isoforms.
Updated Expression Navigator
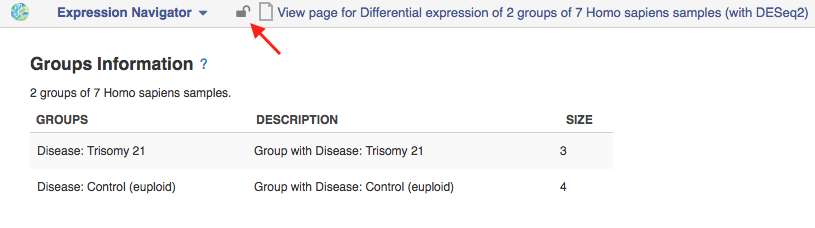
The Expression Navigator is an app that allows users to seamlessly sieve through millions of variants and apply various filters to find the desired results. A new cool feature of the Expression Navigator, is that now you can lock the file in the desired state, freezing the results.
We hope you enjoy using the new apps! As always, the release also comes with bug fixes and performance enhancements. To read our previous release, go
here. Stay tuned for more Genestack news! If you'd like to get in touch with us, please drop us a line at
support@genestack.com 

 We hope you enjoy using the new apps! As always, the release also comes with bug fixes and performance enhancements. To read our previous release, go here. Stay tuned for more Genestack news! If you'd like to get in touch with us, please drop us a line at support@genestack.com
We hope you enjoy using the new apps! As always, the release also comes with bug fixes and performance enhancements. To read our previous release, go here. Stay tuned for more Genestack news! If you'd like to get in touch with us, please drop us a line at support@genestack.com