Last month we rolled out an update to the platform, with several important new features.
New applications: command line tool integration It is easy to integrate command line based tools, like bowtie, bwa, or custom scripts into Genestack. Command Line Applications (CLAs) are a uniform, flexible mechanism to make command line utilities available as Genestack Applications. This update includes the following CLAs:
- Sequencing Aligner - TopHat integrates TopHat, a mapper for RNA-seq reads.
- Bisulfide Sequence Mapping - integrates bsmap, a mapper for bisulfite sequencing reads.
- Methratio - integrates the bsmap methratio script, which computes and filters methylation ratios after mapping.
- HTSeq count - integrates the htseq-count utility from HTSeq, for calculating read counts from RNA-seq read mapping.
- Normalization - integrates our own script for compute RPKM and library-size normalized expression values from read counts.
All CLAs have a common user interface and behavior:
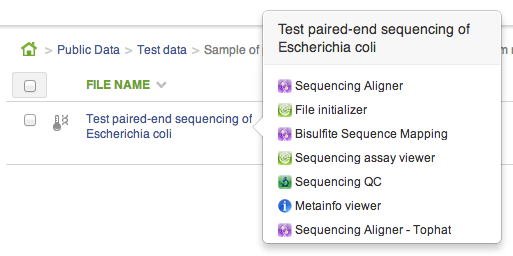
CLAs are used to create new and open existing files. Whenever you click on a file in the file browser, if a CLA can be used to create a file from the one you clicked, it will show up in the context menu. As soon as you open the CLA, the new file will be created:
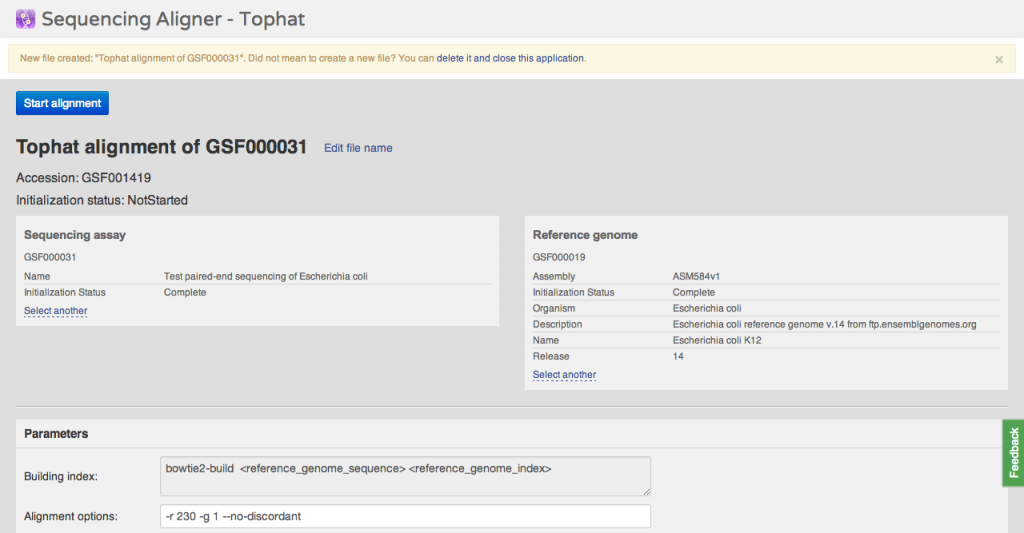
You can immediately undo this action and delete the new file. Alternatively, you can change various parameters: source files and command line arguments. Files are created in
uninitialized state, which means they occupy almost no space, can be used as sources for other applications. You can initialize them later on, when you need the computed data.
Task Manager A simple, light-weight task viewer application is available by clicking the "Tasks" link in the top-right corner of any Genestack Platform screen. It shows recently run tasks in the system together with their statuses and standard and error logs, if available. Note that all computational tasks are file initialization tasks, i.e. directly associated to Genestack files.
Optimizations We improved significantly the speed of autocomplete and BED track visualizations in the Genome Browser. We also added a new, easier to use file chooser component to the Genome Browser and CLAs. We also made it possible to restore forgotten passwords. Happy computing!
 CLAs are used to create new and open existing files. Whenever you click on a file in the file browser, if a CLA can be used to create a file from the one you clicked, it will show up in the context menu. As soon as you open the CLA, the new file will be created:
CLAs are used to create new and open existing files. Whenever you click on a file in the file browser, if a CLA can be used to create a file from the one you clicked, it will show up in the context menu. As soon as you open the CLA, the new file will be created:  You can immediately undo this action and delete the new file. Alternatively, you can change various parameters: source files and command line arguments. Files are created in uninitialized state, which means they occupy almost no space, can be used as sources for other applications. You can initialize them later on, when you need the computed data. Task Manager A simple, light-weight task viewer application is available by clicking the "Tasks" link in the top-right corner of any Genestack Platform screen. It shows recently run tasks in the system together with their statuses and standard and error logs, if available. Note that all computational tasks are file initialization tasks, i.e. directly associated to Genestack files. Optimizations We improved significantly the speed of autocomplete and BED track visualizations in the Genome Browser. We also added a new, easier to use file chooser component to the Genome Browser and CLAs. We also made it possible to restore forgotten passwords. Happy computing!
You can immediately undo this action and delete the new file. Alternatively, you can change various parameters: source files and command line arguments. Files are created in uninitialized state, which means they occupy almost no space, can be used as sources for other applications. You can initialize them later on, when you need the computed data. Task Manager A simple, light-weight task viewer application is available by clicking the "Tasks" link in the top-right corner of any Genestack Platform screen. It shows recently run tasks in the system together with their statuses and standard and error logs, if available. Note that all computational tasks are file initialization tasks, i.e. directly associated to Genestack files. Optimizations We improved significantly the speed of autocomplete and BED track visualizations in the Genome Browser. We also added a new, easier to use file chooser component to the Genome Browser and CLAs. We also made it possible to restore forgotten passwords. Happy computing!