Genestack attended Illumina's DNA Technology Symposium, a celebration for reaching the "1000 dollar genome" milestone. The event was kicked off by Prof. Shankar Balasubramanian, co-founder of Solexa, whose collaboration with David Klenerman led to the discovery of the sequencing-by-synthesis technology that made Illumina leader of the sequencing market.
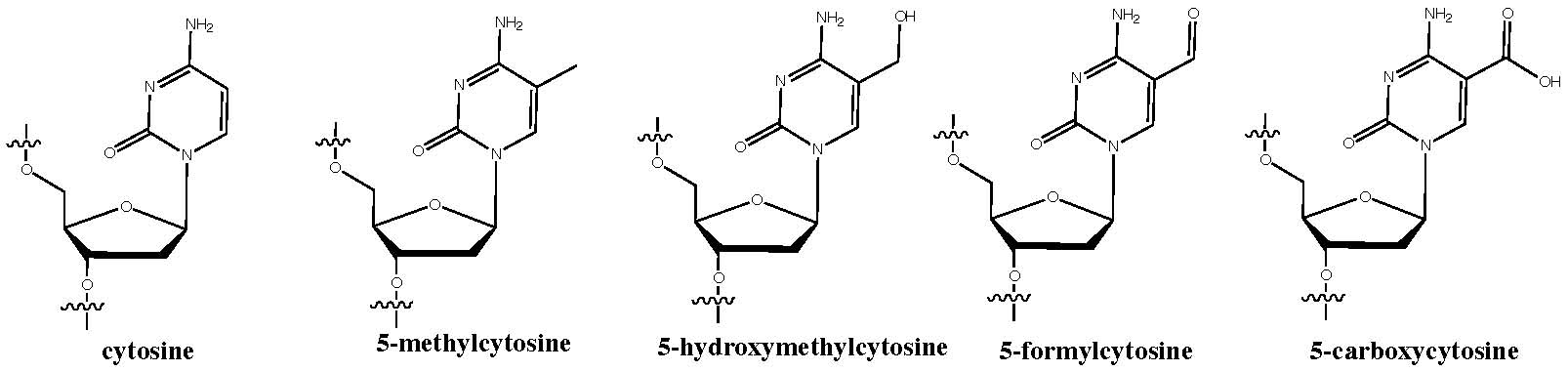
Prof. Balasubramanian took the audience beyond the usual four bases, discussing his group's work about Reduced Bisulfite sequencing (redBS-seq) and Oxidative Bisulfite sequencing (oxBS-Seq). The combination of these methods allows discriminating between 5-formylcytosine (5fC), 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) modifications, albeit at the cost of twice the sequencing depth.
Joe Boutell from Illumina San Diego took us down History lane, showing the evolution of Illumina's clustering technology, reminding us of the good old times "when we were still competing with SOLiD and Roche". Tim Albrecht from Imperial College London made clusters even more historical, introducing us to recent advances in tunnelling-based sequencing.Similarly to Oxford Nanopore technology, this third-generation sequencing method not only works on single DNA or RNA molecules, but has the potential to be applied to peptides. However, Dr. Albrecht stressed how recents attempts at peptide sequencing required nothing less than 52 signal features, cooling our enthusiasm.
Prof. Tom Brown from Oxford delighted us with another nanotechnological application, an artificial method of polymerisation and ligation of DNA based on click chemistry. The more natural side of sequencing was well represented by several talks about DNA repair, DNA replication fidelity and mutant polymerases.
Lyn Chitty from NHS gave one of the most interesting talks, discussing the NHS's first results using non-invasive prenatal testing (NIPT) based on sequencing cell-free fetal DNA, reminding us the fundamental role of sequencing for the future of public healthcare.
Overall, it's been a day full of exciting talks, but something was missing. It is easy to share the enthusiasm for being able to sequence a whole genome in 29 hours for 1000 USD using ten machines, but that was only part of the story. It struck us the hardest at the reminder of Illumina representing "90% of the entire world's sequencing data". It is a bizarre contradiction: today sequencing data might be cheaper to make, but it's increasingly expensive to use. In other words, today it takes much more than 1000 dollars to "unlock the power of the genome".
First, any amount of data is useless if you don't have access to it. The difficulty of accessing genLife Science Datasets is a known problem: the presence of political and technical barriers is a major limitation to clinical research, and it would have been great to hear more about Illumina's initiatives for promoting open access to sequencing data.
Second, any amount of reads is nothing more than lorem ipsum if you can't analyse it. More than a decade later, researchers are still wasting their limited time in a swamp of file formats, single-threaded Perl scripts and makefile shenanigans. Illumina might have biochemistry and enzymology as the bases of its success, but it would have no backbone without Bioinformatics. The truly "unlock the power of the genome" we must empower the scientific community with the tools and resources required to handle a rising ocean of sequencing data. Luckily for Illumina and for its customers, Genestack is working hard to allow researchers and clinicians to make the most of next-generation sequencing data. Genestack's format-free files and modular applications are designed to help users focus on analysis and interpretation, putting Biology first. Moreover, our Data Flow system allows users to easily share workflows and pipelines, enabling fully reproducible research. In parallel, Genestack's Public Data repository offers free, direct access to thousands of public experiments and assays, helping researcher making the most of datasets that have already been shared and promoting open access.
Illumina might have biochemistry and enzymology as the bases of its success, but it would have no backbone without Bioinformatics. The truly "unlock the power of the genome" we must empower the scientific community with the tools and resources required to handle a rising ocean of sequencing data. Luckily for Illumina and for its customers, Genestack is working hard to allow researchers and clinicians to make the most of next-generation sequencing data. Genestack's format-free files and modular applications are designed to help users focus on analysis and interpretation, putting Biology first. Moreover, our Data Flow system allows users to easily share workflows and pipelines, enabling fully reproducible research. In parallel, Genestack's Public Data repository offers free, direct access to thousands of public experiments and assays, helping researcher making the most of datasets that have already been shared and promoting open access. Thanks to Illumina, today mankind has unprecedented access to its own blueprint. Genestack's team looks forward to new, exciting updates from Illumina's technology: meanwhile, we'll make sure you can make the most of it.