Introduction
At a first glance, this task may appear daunting: storing proteomics data and blood samples from a population of 400,000 individuals in a single system, with the added requirement of retrieving the data directly from the Jupyter notebook without even opening the system's interface. It sounds impossible, but in this paper we’ll do just that; as we analyze the proteomics of atopic dermatitis and see how easy this impossible task can be with one tool - Genestack's Open Data Manager (ODM).
This study highlights how our data management platform optimises proteomic analysis with publicly available data. With Genestack’s ODM, researchers can enable true proteomic analysis by merging low and high complexity data into a single platform. In this article, our objective is to identify proteins involved in hBD-2-mediated protection of the human skin barrier in vitro. This study will also compare the results to blood samples of patients with atopic dermatitis to validate the results.
Genestack’s Open Data Manager For Proteomics
The analysis of proteomics data presents researchers with several critical challenges. These challenges include selecting relevant expression values based on metadata, cleaning data efficiently and working with multiple datasets. All these problems can be solved simply with the use of ODM, as it helps to query signal data based on metadata, work with several different datasets simultaneously and does not require more than a single afternoon for a thorough proteomics analysis with validation of results using blood samples. It really can be that easy.
Proteomics analysis can also be complex, as data is usually stored in different formats and requires a lot of preprocessing before analysis can be started. In a system that is built specifically for biological data, including proteomics, complex queries can be executed with ease, hence providing a quick and efficient analysis of the data. There is no one-size-fits-all solution for data storage as OMICS data is extremely different from other types of data, such as marketing or financial data and requires a very distinct approach. Therefore, the importance of employing a specialised solution to meet the unique requirements of biological data management cannot be overstated.
Cleaning proteomic data can be a tedious and time-consuming process. The Genestack platform's API enables the loading, selection, and analysis of proteomic data programmatically, resulting in significant time and effort savings. We believe that scientists should spend their time positively impacting people's lives as opposed to being bogged down with the tedious task of sifting through and harmonizing data.
Working with multiple datasets is a significant challenge due to variations in sample preparation, experimental design, and data formats. However, the Genestack platform can assist in unifying and integrating multiple datasets using data templates and scientific ontologies, and allows you to upload all your scientific datasets to one platform; thus enabling cross-study comparisons and analyzes. Furthermore, the platform's dictionary feature can be utilised to unify the terms used in different datasets, allowing for efficient data analysis. Thus, the Genestack platform provides a comprehensive solution for the efficient analysis of proteomics data, addressing the challenges associated with handling and interpreting large and complex datasets.
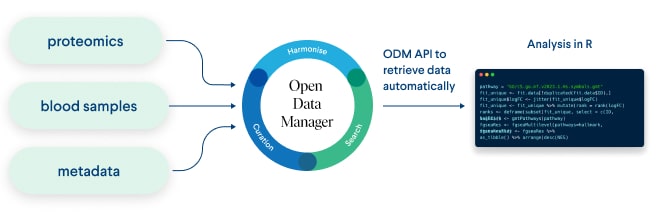 Figure 1. Schema of data flow from external sources to ODM and then to analysis tools using API
Figure 1. Schema of data flow from external sources to ODM and then to analysis tools using API
Materials and Methods
The Genestack platform served as the primary tool for data storage, handling and management for this study. The platform's robust application programming interface (API), coupled with its dedicated R packages, enabled the seamless loading, cleansing and retrieval of data. The Genestack platform's state-of-the-art features were leveraged to facilitate efficient data handling and streamline the analysis process.
Proteomic data was collected from the ProteomeXchange1 and the PRIDE2 databases. Statistical methods such as t-test and ANOVA were utilised to identify differentially expressed proteins. Functional enrichment analysis using gene ontology pathway analysis was employed to gain insight into biological processes associated with differentially expressed proteins. Candidate biomarkers were validated using blood samples of atopic dermatitis patients from the UK Biobank3 that were loaded directly to ODM along with proteomics data. Finally, the R programming language with packages such as limma4, for differential expression analysis and fgsea5 for GSEA analysis was used to process and visualize the data.
Analysis and Interpretation
The initial phase of the analysis entailed exploratory data examination, which uncovered a total of 2051 unique proteins. Upon further inspection, it was noted that the control condition exhibited the highest number of proteins, whereas the lowest number of proteins was detected in samples treated with V8 toxin, which is a relative of atopic dermatitis. This preliminary analysis provided a useful starting point and enabled us to develop a deeper understanding of the distribution of proteins across different conditions.
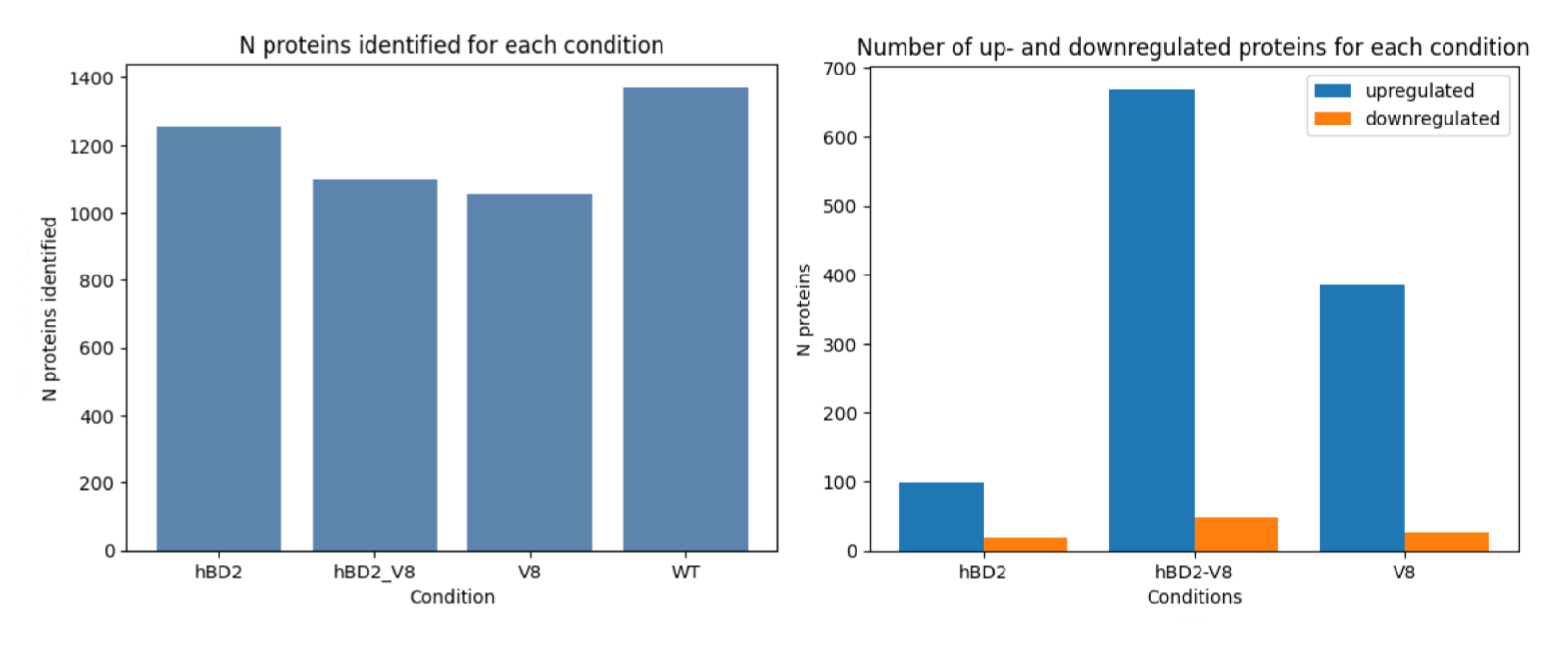 Figure 2. Barplots of number of proteins identified in each state and number of up- and down regulated proteins compared to control state (WT)
Figure 2. Barplots of number of proteins identified in each state and number of up- and down regulated proteins compared to control state (WT)
Subsequently, a differential expression analysis was conducted, wherein all three states were compared to the control. The results indicated that the highest number of upregulated proteins was observed in samples treated with both the V8 toxin and human beta-defensin peptide, thereby indicating a significant change compared to samples treated solely with the toxin. These findings suggest that human beta-defensin 2 (hBD2) does not typically elicit a substantial response in healthy skin but does so in skin treated with the toxin, which was proved by research6. This observation serves as a promising indication that hBD2 may prove to be an effective treatment option for atopic dermatitis.
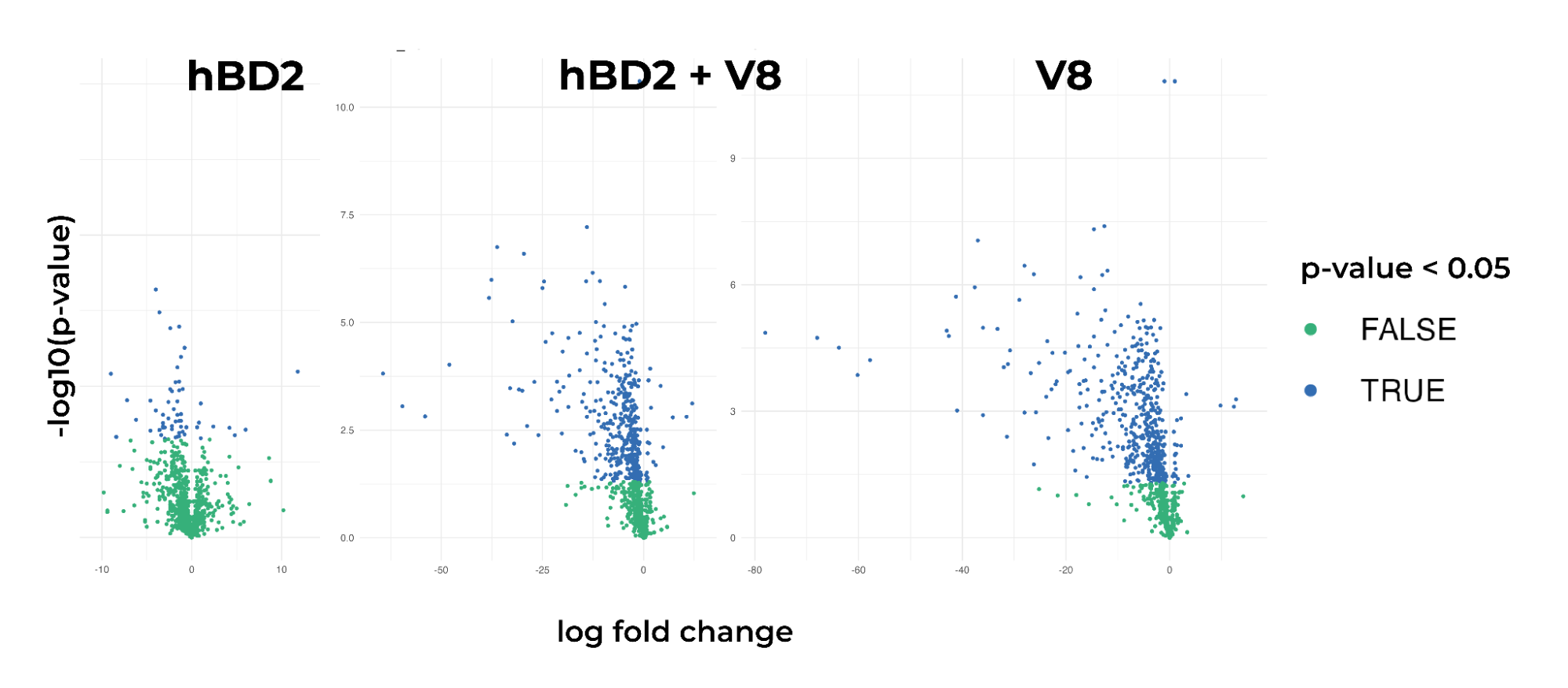 Figure 3. The volcano plot of diff expressed proteins in three different states
Figure 3. The volcano plot of diff expressed proteins in three different states
Furthermore, volcano plots were generated to visually compare the three states. They revealed a stronger response in samples treated with V8 or V8 + hBD2 compared to hBD2 alone. These findings suggest that hBD2 may not be the best candidate for protecting healthy skin but could be a useful component for treating atopic dermatitis.
In the third phase of our analysis, a more targeted approach was adopted to investigate the molecular pathways under different conditions. The results revealed that in the V8 state, most of the pathways were downregulated, which provides evidence for the toxic nature of V8. However, some pathways showed significant overexpression (p-value < 0.01), including such pathways as DNA replication and collagen receptor binding. Apolipoprotein emerged as a potential biomarker, as the apolipoprotein binding pathway was significantly enriched in skin samples treated with V8 toxin. Interestingly, this pathway was also enriched in the V8 + hBD2 treatment, suggesting that it may serve as a promising biomarker for atopic dermatitis.
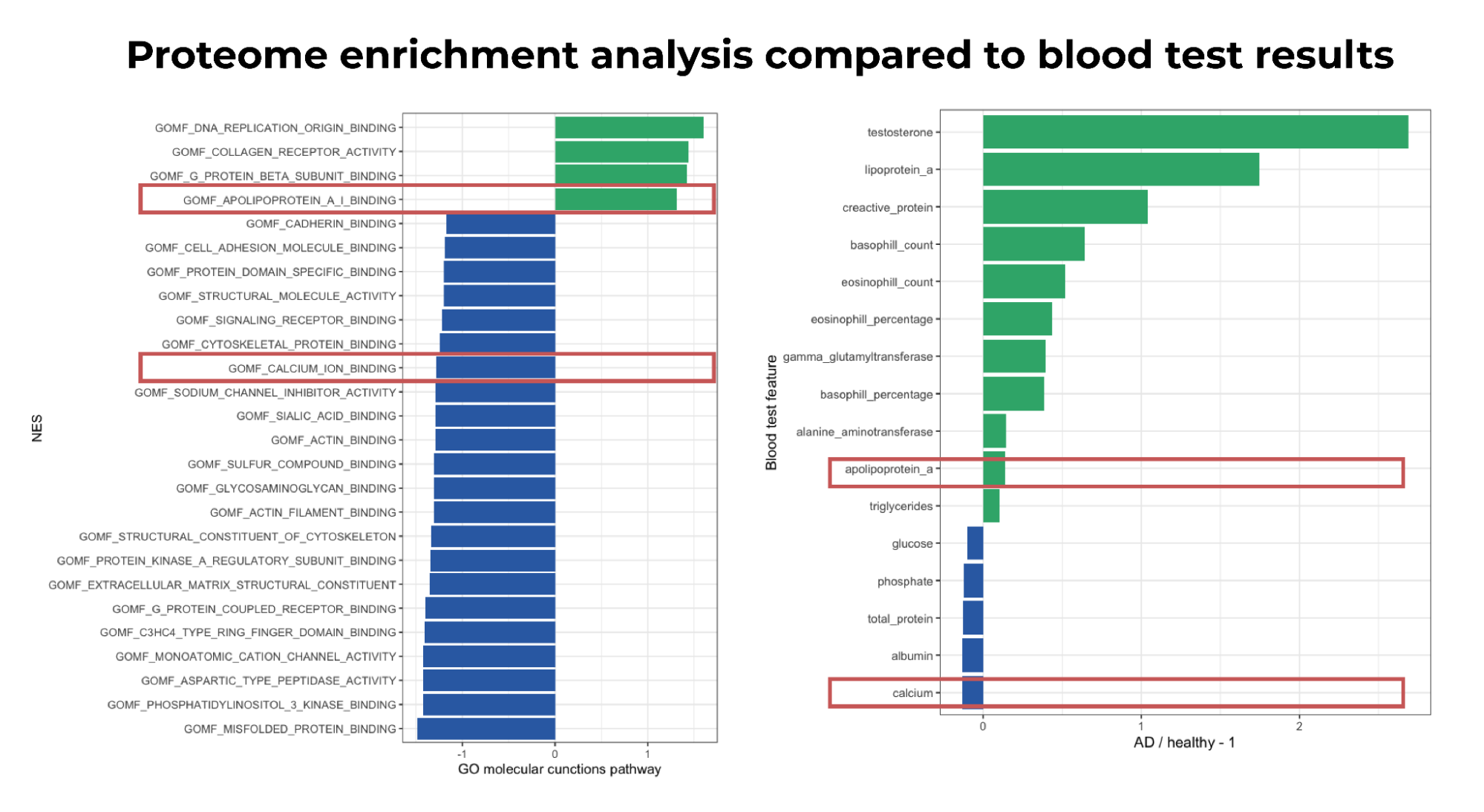 Figure 4. The barplots of Go molecular functions pathway in V8 state and blood samples of atopic dermatitis patients compared to healthy individuals. Highlighted are features that show similar patterns
Figure 4. The barplots of Go molecular functions pathway in V8 state and blood samples of atopic dermatitis patients compared to healthy individuals. Highlighted are features that show similar patterns
Finally, we sought to validate our results using external data from the UK Biobank 3. Specifically, we analyzed 375,677 blood samples, including 11,253 samples from individuals with atopic dermatitis. Using one-way ANOVA, we compared the blood test results of atopic dermatitis patients to those of healthy individuals. The results revealed that several blood test features were elevated in atopic dermatitis patients compared to healthy individuals, including C-reactive protein, which has earlier been identified as a biomarker of different skin conditions7. Apolipoprotein was also elevated, which was previously identified as enriched in V8-treated skin samples. The role of apolipoprotein in skin health has also been proven in research8. Additionally, the calcium ion binding pathway was downregulated, aligning with the decreased calcium levels observed in the blood samples of atopic dermatitis patients, further supporting our earlier findings.
Conclusion
The aim of the study was to identify signalling pathways involved in hBD-2-mediated human skin barrier protection in vitro using a publicly available proteomics dataset. To achieve this, Genestack’s ODM platform was used to consolidate proteomics data and low-throughput data in one place. This approach allowed for a comprehensive analysis of the data, which was completed within one afternoon. Using ODM streamlined the proteomics analysis and allowed most of the time to be spent on insightful analysis rather than data cleaning and searching. This highlights the potential of the Genestack platform to improve the efficiency and effectiveness of proteomics research.
For those interested in exploring the platform, Genestack offers a live demonstration and a free trial. Contact us to learn more:
info@genestack.com
https://genestack.com/products/omics-data-manager/
Literature
- https://www.proteomexchange.org
- https://www.ebi.ac.uk/pride
- https://www.ukbiobank.ac.uk
- https://bioconductor.org/packages/release/bioc/html/limma.html
- https://rdrr.io/bioc/fgsea/man/
- https://www.nature.com/articles/s41598-023-29558-0
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4513404/
- https://www.sciencedirect.com/science/article/pii/S0022202X15338471
