Introduction
The integration of diverse data types in biomedical research is becoming increasingly important as scientists strive to develop a more holistic understanding of complex biological systems. One area where this integration is particularly valuable is in the study of brain activity through Electroencephalography (EEG). Traditionally used in clinical and research settings to monitor electrical activity in the brain, EEG data offers insights into neurological conditions, cognitive processes, and brain function.
Yet traditional neuroinformatics workflows can take 6–12 months to correlate EEG patterns with genomic data for treatment decisions, a delay that can slow patient care and research breakthroughs. Integrated platforms can cut this to days, enabling:
- Faster time-to-treatment for personalized neurological care
- Reduction in duplicated research efforts across institutions
- Annual savings industry-wide from eliminated data silos
Genestack's Open Data Manager (ODM) has been a cornerstone for managing Life Science research data, but its capabilities extend beyond traditional bioinformatics applications. This case study explores how EEG data in European Data Format (EDF) can be integrated into ODM alongside other omics datasets. The result: a versatile, unified platform for neuroinformatics research with far-reaching possibilities.
Understanding EEG and EDF
EEG is a technique that measures electrical activity in the brain using electrodes placed on the scalp. It is widely used to diagnose and monitor neurological disorders such as epilepsy, sleep disorders, and brain injuries, where rapid and accurate data analysis can directly impact treatment decisions. The data recorded during EEG sessions are typically stored in EDF, a standardized format designed for storing multichannel bioelectrical signals. EDF files not only contain the raw signal data but also include essential metadata such as sampling rates, electrode locations, and patient demographics.
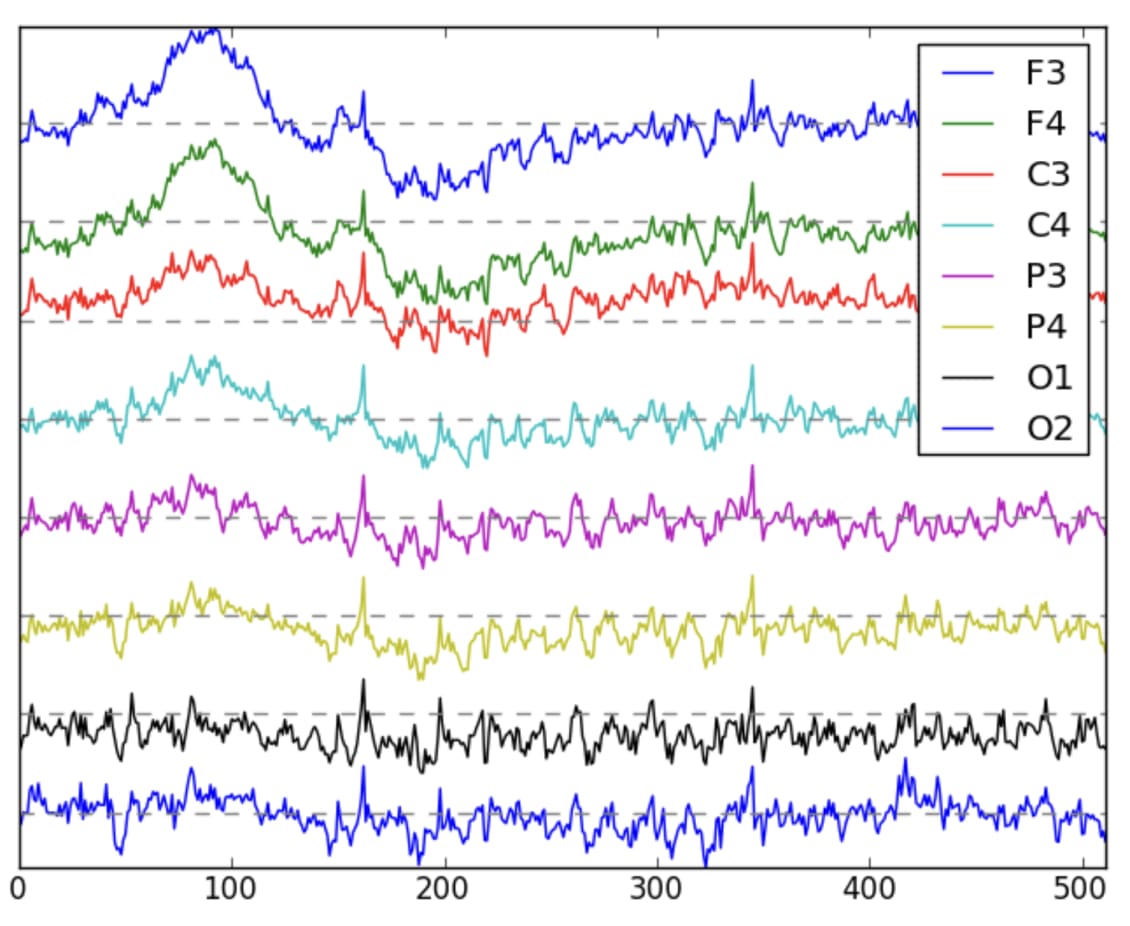
Example of EEG data visualized [1]
Beyond technical compatibility, this EDF integration capability solves a critical business problem: data accessibility bottlenecks. Hospitals generate terabytes of EEG data monthly, but remains isolated in proprietary systems which in turn slows down research collaboration and delays patient care.
The Challenge
Managing EEG data presents several unique challenges:
- Data Volume and Complexity: EEG studies can produce vast amounts of data, especially when recorded over extended periods or across multiple channels. Handling this volume of data requires efficient storage solutions and powerful computational tools.
- Metadata Management: Accurate and comprehensive metadata is essential for contextualizing EEG data, facilitating data sharing, and ensuring the reproducibility of research findings.
- Integration with Other Data Types: For a comprehensive understanding of brain function, EEG data often needs to be integrated with other data types, such as genetic, imaging, and clinical data. This integration requires a flexible and interoperable data management platform.
- Security and confidentiality: Security and confidentiality are crucial for EEG medical data research because this data often contains sensitive personal health information, which must be protected to comply with privacy laws and regulations. Additionally, maintaining confidentiality is essential for the ethical conduct of research and ensuring that participants' identities and medical histories are safeguarded.
- Ease of Use: If the platform used to manage this data is not user-friendly, it can lead to inefficiencies, errors, and frustration, especially for researchers who may not be highly technical. A steep learning curve or cumbersome interface can slow down the research process, making it difficult to perform tasks like data cleaning, annotation, or visualization effectively.
Why ODM?
Genestack's ODM is ideally suited to address these challenges, providing a robust infrastructure for managing and analyzing complex datasets. Here are some key features of ODM that make it an excellent choice for EEG data management:
- Scalable Storage and Organization: ODM offers scalable solutions that can accommodate the large volumes of data generated by EEG studies. Its flexible data organization capabilities allow researchers to categorize and manage data effectively, ensuring that it remains accessible and useful for future analyses.
- Comprehensive Metadata Support: ODM's metadata management capabilities are particularly valuable for EEG data. The platform allows for the detailed annotation of datasets, capturing information about the experimental setup, data collection parameters, and patient characteristics. This metadata is critical for data interpretation, quality control, and facilitating secondary data use. This metadata standardization eliminates the manual curation bottlenecks that typically consume 30-40% of researchers' time. ODM also ensures consistent adherence to agreed vocabularies and ontologies to ensure that data is consistent and findable.
- Data Integration and Interoperability: ODM supports the integration of EEG data with other types of biological and clinical data, enabling researchers to conduct multidisciplinary studies. The platform's data model can accommodate diverse data formats and standards, facilitating seamless data integration and analysis.
- Security and Confidentiality: Genestack’s ODM product is developed through establishing requirements that have been cross-referenced and aligned to 21CFR Part 11, ICH Guideline for good clinical practice E6 (R2) and ISO 27002 information security controls. These requirements are a primary input to the design and development of ODM and enable our clients to comply with the applicable regulatory requirements during the use of the product. [2]. This compliance-by-design approach helps clients meet regulatory requirements during use, eliminating delays of 6–12 months in multi-institutional approvals.
- User friendly: ODM is designed with the user in mind, bringing the possibilities of organizing, curating and retrieving data via both GUI, Python scripts and APIs, depending on your technical expertise. These APIs enable the kind of real-time data access that transforms research from batch processing to continuous discovery workflows
- Return on Investment Reality: Organizations typically spend 18-24 months and $2-5 million building internal neuroinformatics integration capabilities. ODM eliminates this infrastructure burden, allowing research teams to focus on discovery rather than data engineering. Early adopters report significantly faster completion of multi-modal studies, moving from data wrangling to discovery in a fraction of the usual time.
Case Example: Uploading and Utilizing EEG Data in ODM
EEG data in EDF format is inherently complex, often comprising raw signals, annotations, and header information within a single file. This complexity presents significant challenges for researchers in terms of storage, retrieval, and organization of their studies. In this case study, we explore how EEG data in EDF format can be effectively stored in ODM and discuss the necessary preprocessing steps to optimize the user experience.
For this example, we utilize two EDF files from the study “EEG Signals from an RSVP Task” by Matran et al. (2017) [3], available on the PhysioNet open database.
These selected EDF files were generated using the Rapid Serial Visual Presentation (RSVP) protocol, where images of aerial London were presented at a speed of 5 Hz. The EEG signals were captured from eight channels according to the 10-20 system (PO8, PO7, PO3, PO4, P7, P8, O1, and O2) and are named using the format `rsvp_5Hz_{participant_id}{a,b}.edf`. The suffix (either "a" or "b") indicates the sequence in which the files were recorded. Each file contains continuous EEG recordings sampled at 2048 Hz, with signals filtered between 0.15 and 28 Hz. The data is measured in microvolts.
Data Preparation and Organisation
Deconstruction of EDF file
The first step involves deconstructing the EDF file to gain a deeper understanding of the contained data. We utilize Python 3 and the Jupyter Notebook framework for this task, leveraging the `pyedflib` library, which provides an efficient and straightforward way to handle EDF files. By using `pyedflib`, we can easily extract three DataFrames from an EDF file: `raw_signal`, `header`, and `annotations`.
Alignment with the ODM data model
With the raw data extracted, the next step is to align it with the ODM data model. The following actions are taken to prepare the data:
- Column Renaming: Adjusting column names to facilitate easy connection with participant IDs.
- Adding Sample Source ID: Introducing a primary connector between the files.
- Creating a Unified Header: Combining each header file with additional participant data to form a comprehensive header file, capturing all relevant participant information.
- Adding Attributes: Incorporating additional attributes into the header file.
- File Conversion: Converting the data to TSV format, ensuring compatibility for upload to ODM.
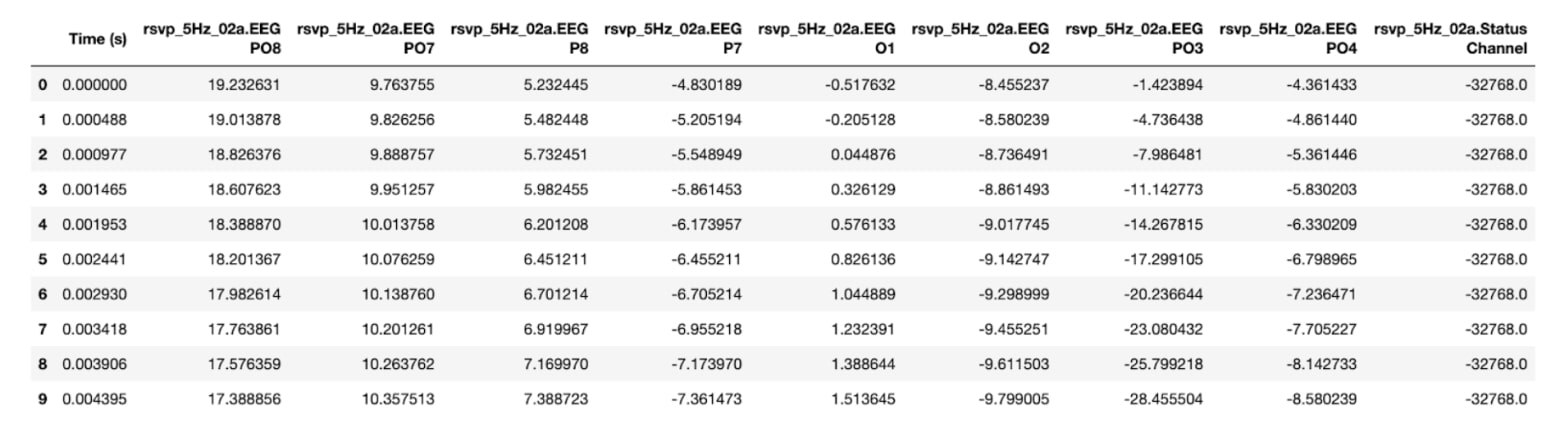
Figure 2. Raw signal file (first 10 rows; rsvp_5Hz_02a) - saved as rsvp_5Hz_02a.tsv
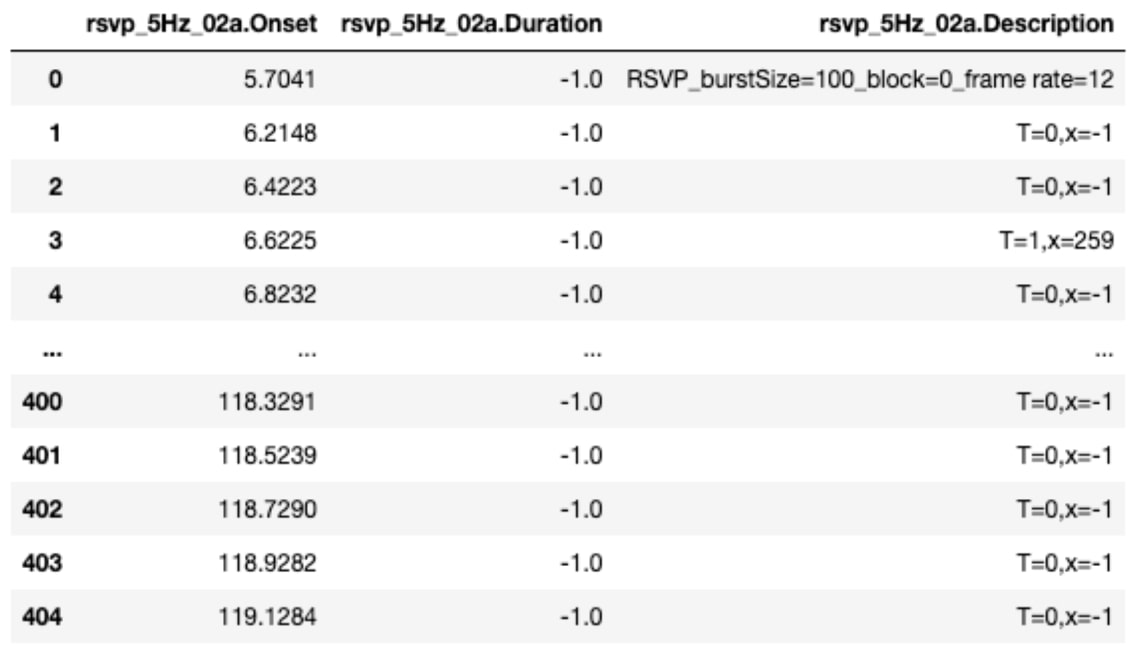
Figure 3. Annotations (rsvp_5Hz_02a) - saved as rsvp_5Hz_02a.tsv
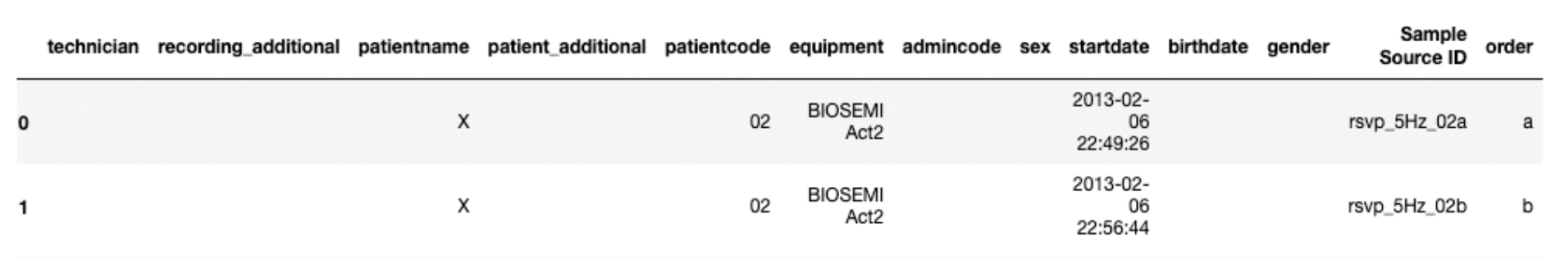
Figure 4. Header (Samples data with rsvp_5Hz_02a and rsvp_5Hz_02b) - saved as rsvp_5Hz_02.tsv
Data Upload
Having prepared files, it is easy to upload them to ODM by using the graphical user interface. Following the steps presented, simply create a new study, choose a template of your choice, and upload samples’ metadata (file saved as *rsvp_5Hz_02.tsv*).
Subsequently, in the "Data" section, upload the remaining four data files (two raw signals and two annotations - *rsvp_5Hz_02a_annotations.tsv, rsvp_5Hz_02b_annotations.tsv, rsvp_5Hz_02a.tsv, rsvp_5Hz_02b.tsv*), specifying the delimiter as `"."`. Now, you have your data ready for further analysis, curation or visualization.
Integrative Approach to Omics and Imaging Data
In Genestack's Open Data Manager (ODM), researchers can seamlessly combine omics data—such as genomics, transcriptomics, and proteomics—with brain imaging data like EEG, MRI, PET. This integration is crucial for advancing our understanding of brain diseases because it enables a comprehensive analysis of both molecular and imaging data within a single platform. By unifying these diverse data types in ODM, researchers can uncover connections between molecular pathways and brain structure or function, that are often missed when analyzed separately. This holistic approach accelerates the discovery of novel therapeutic targets and biomarkers, facilitating the development of more precise and effective treatments for brain diseases. Additionally, ODM’s user-friendly interface ensures that even complex datasets can be managed and analyzed efficiently, making it an invaluable tool for pioneering new discoveries in brain research. [3]. This ensures researchers can focus on insight generation rather than data wrangling.
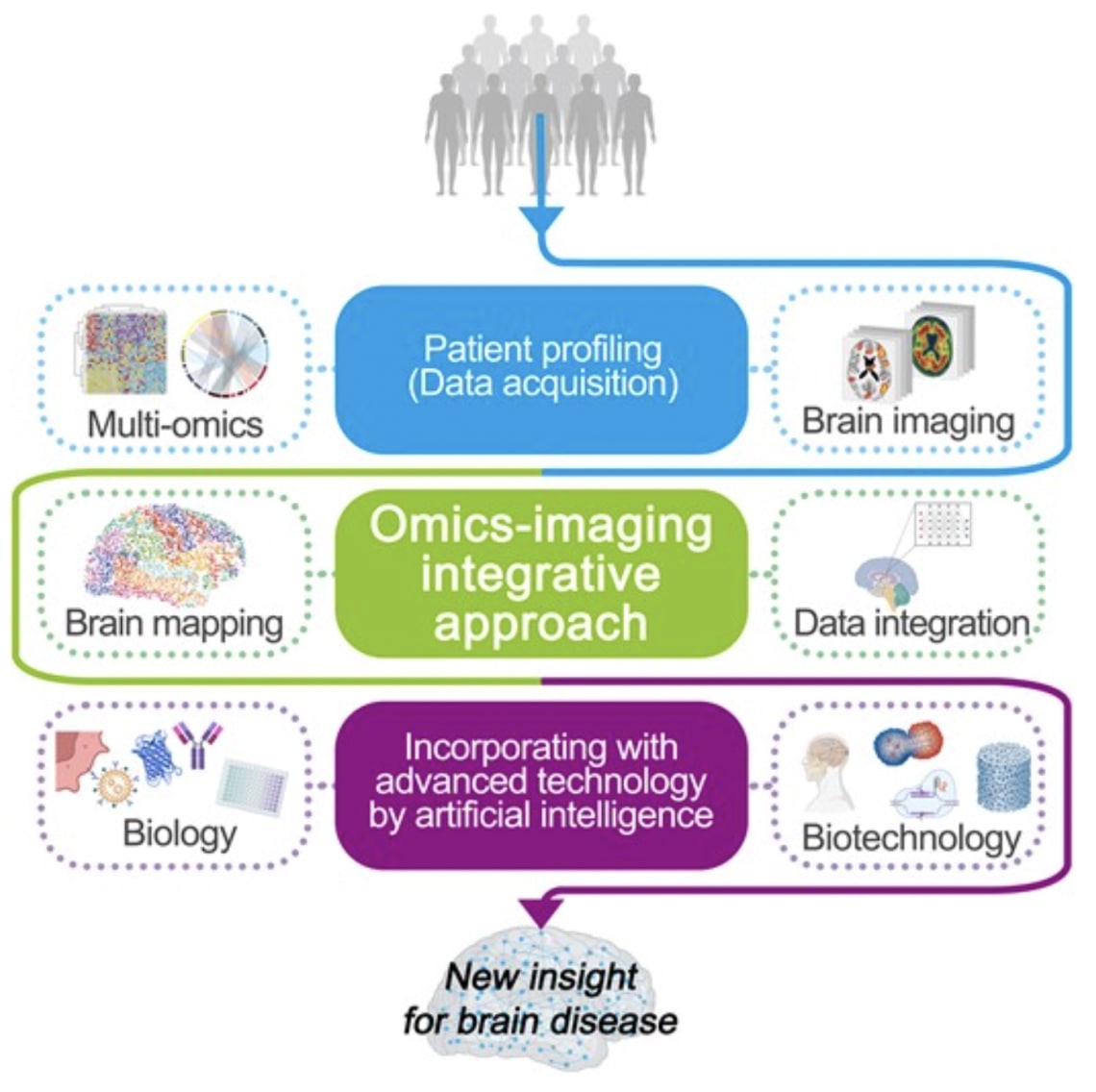
Figure 5. Integrative approach of omics and imaging data to discover new insights for understanding brain diseases [3]
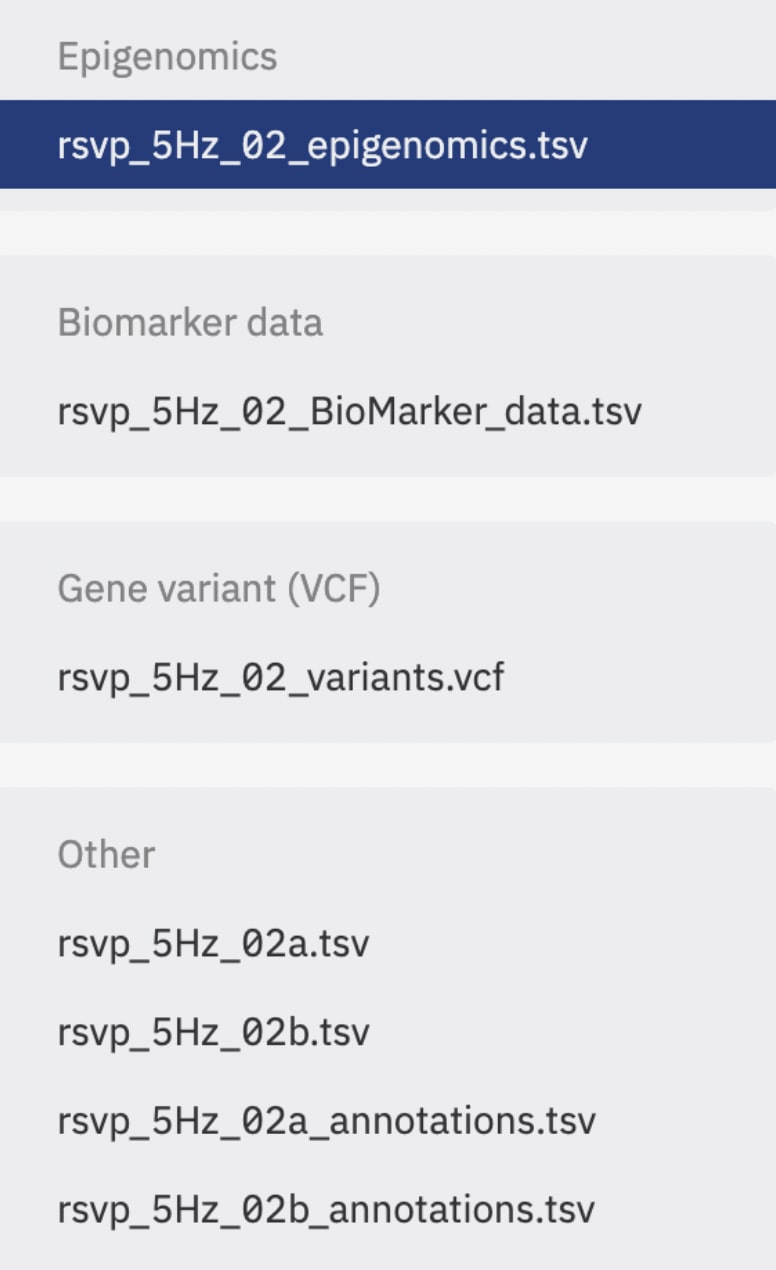
Figure 6. Example of variants, biomarker and epigenomics data, combined with EEG signal and annotations data stored in ODM.
Disclaimer: Epigenomics, Biomarker and Gene variant data is example data and not connected to study “EEG Signals from an RSVP Task” by Matran et al. (2017).
Data Analysis and Visualization
Once the data is housed in ODM, researchers can proceed with analysis using ODM's curation capabilities. For more advanced analyses, ODM supports integration with external tools and software (shown below EEGLAB software as an example), enabling the application of specialized signal processing techniques or machine learning algorithms. This flexibility is essential for addressing the diverse analytical requirements of neuroinformatics research. Furthermore, integrating the output data back into ODM ensures that all research data is centralized, organized, and accessible, streamlining the research process.
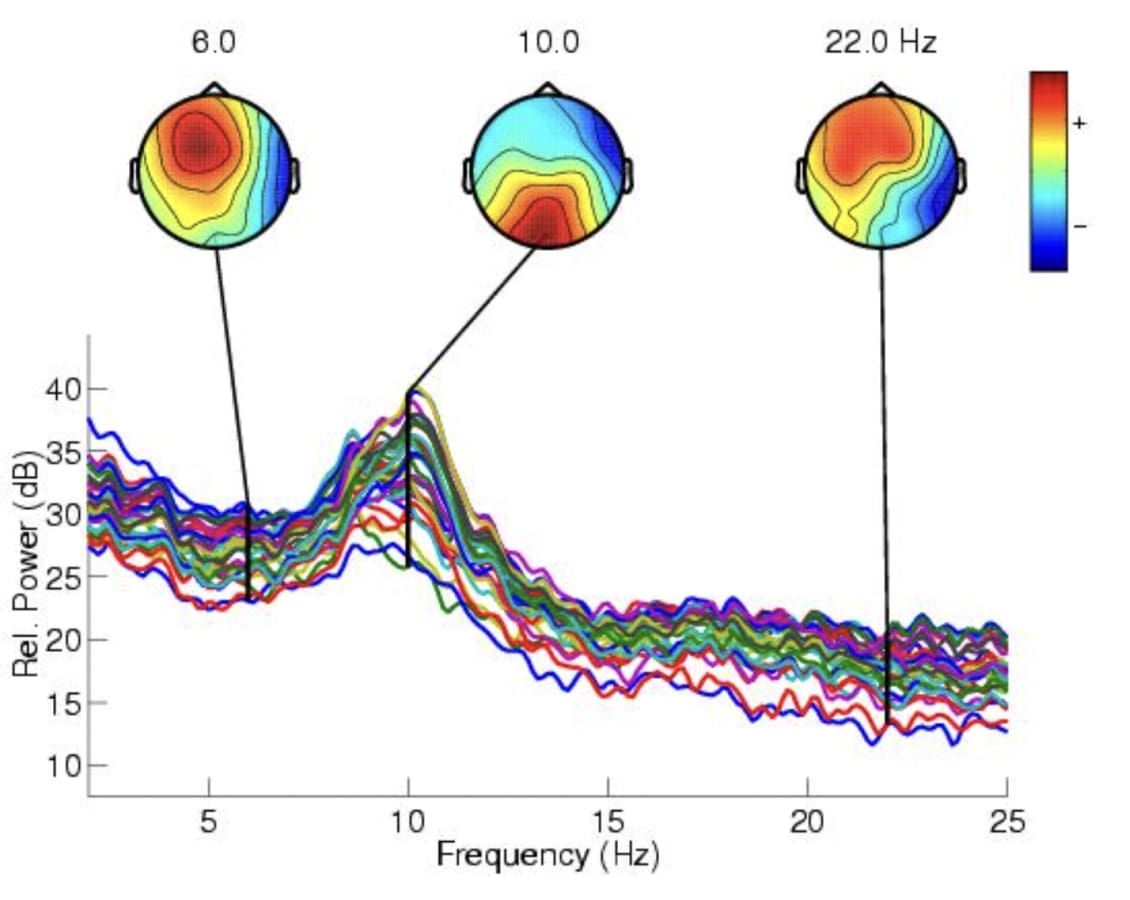
Figure 7. Example of Power Spectrum and Scalp Maps generated by EEGLAB toolbox [5]
Collaboration and Data Sharing
One of the strengths of ODM is its support for collaborative research. The platform allows researchers to share datasets, analysis workflows, and results with collaborators. This feature is particularly valuable for large-scale studies that involve multiple research groups or institutions, as it facilitates data sharing, coordination, and reproducibility.
Conclusion
As the article has shown, in the rapidly evolving field of neuroinformatics, integrating diverse datasets is key to unlocking new insights into brain function and disorders. We have demonstrated how Genestack's Open Data Manager (ODM) can streamline the management of EEG data in EDF format, combining it seamlessly with other omics data to create a holistic view of brain activity. By leveraging ODM's robust data management capabilities, researchers can overcome common challenges associated with large-scale EEG data, including data volume, metadata management, and integration with other biological data types.
The ability to integrate EEG data with genomics, transcriptomics, proteomics, and other data types within ODM not only enhances data accessibility and organization but also facilitates comprehensive analyses. This synergy accelerates the discovery of novel biomarkers and therapeutic targets, ultimately advancing our understanding of brain diseases.
Moreover, ODM's compatibility with external tools extends its analytical power, offering researchers the flexibility to apply specialized techniques and algorithms. This integration ensures that complex data analyses can be conducted efficiently, while maintaining a centralized and organized data repository.
The platform’s collaborative features further enhance its value, allowing researchers to share and coordinate efforts across multiple institutions, thus promoting reproducibility and accelerating scientific progress. Genestack's ODM uniquely positions researchers as a versatile and user-friendly solution for managing and analyzing neuroinformatics data, paving the way for groundbreaking research and innovations in understanding brain function and disease.
The implications of adopting (or delaying) such an infrastructure are significant. The Strategic Reality: Every month of delay in implementing integrated neuroinformatics infrastructure represents lost competitive advantage. While your organization wrestles with data silos, competitors with integrated platforms are:
- Discovering biomarkers faster
- Attracting better research partnerships
- Winning more grant funding through superior data capabilities
- Building AI advantages from comprehensive datasets
The neuroinformatics revolution isn't coming—it's here! The question facing research leaders: Will your organization lead this transformation or follow it?
The difference between months and minutes in neuroinformatics discovery starts with the right infrastructure decision.
Next Steps:
- Assess your current EEG-omics integration gaps
- Calculate the cost of continued data silos
- Evaluate purpose-built vs. internal development approaches
Ready to Transform Your Neuroinformatics Capabilities?
Contact Genestack to see how leading research organizations are accelerating brain science discoveries.
References
- Colorado State University, EEG data tutorial: Getting started. Retrieved September 4, 2024, from www.cs.colostate.edu
- Genestack, Security and compliance. Genestack. Retrieved September 4, 2024, from genestack.com
- Matran-Fernandez, A., Poli, R., Cinel, C., & Sepulveda, F. (2017). Towards the automated localization of targets in rapid image-sifting by collaborative brain-computer interfaces. PLoS ONE, 12(5), e0178498. doi.org
- Jong Hyuk Yoon, Hagyeong Lee, Dayoung Kwon, Dongha Lee, Seulah Lee, Eunji Cho, Jaehoon Kim, Dayea Kim, Integrative approach of omics and imaging data to discover new insights for understanding brain diseases, Brain Communications, Volume 6, Issue 4, 2024, fcae265, doi.org
- Neuroelectrics, EEGLAB. Retrieved September 4, 2024, from www.neuroelectrics.com
