Hello everyone, We're excited to announce our next release - Platform Release 33! It comes with several new important features and interface improvements as well as bug fixes and performance enhancements.
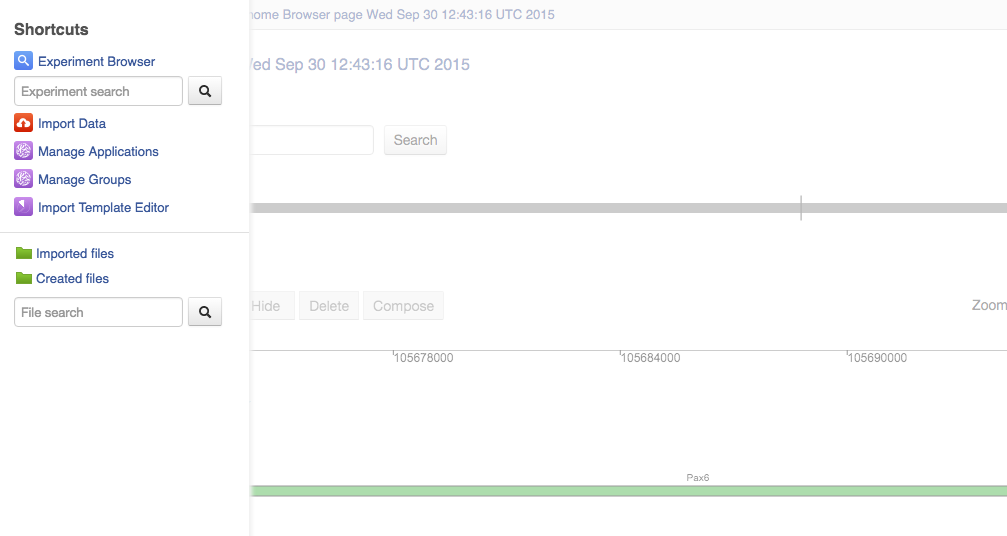
Shortcuts menu
First of all, as you might have noticed, we introduced a shortcuts menu - an easy way to reach the most commonly used apps and folders. Genestack Import, Manage Apps, Manage Groups, Experiment Browser as well as Created and Imported files folders can all be found there. How to access the menu“ Move your mouse cursor to the left hand side of any page. We're sure you'll find this new feature very useful and time-saving.
Login synchronisation
Second important feature we implemented is login synchronisation across the Web Browser tabs after session time-out. Let's say you have a lot of tabs open in your web browser and you step away from your computer - after some time the system will log you out. Previously, you'd have to log in on every one of these pages. Now, you just have to do it in one tab and the others will synchronise.
GenePix microarray support
Our users doing work on microarrays will be happy to hear that recently we implemented support for GenePix microarray assays, on top of already supported Agilent and Affymetrix microarrays. Now when grp.gz data is imported into Genestack, the OS recognizes it as GenePix Microarray assay during import. Later on, users can check the microarray quality with our Microarray QC app, perform microarray normalisation and use the Dose Response Analysis app.
Metainfo templates changes
As you know you can import data into Genestack using Import templates. You can either use a default template or create your own, in which you will specify the required metadata fields, e.g. disease, sex, method. Previously, you would choose the template before data import. After our last update, you can always change this template: after your data is imported and anytime after that. This new option allows our users to change what metainfo fields are required and which are optional. You can also use this option to add new fields to your template or change what ontologies or controlled vocabularies are used with your data. Importantly, these changes will not erase any data already present in your files. How to access this option“ 1) while you're importing data: in the Edit metainfo step, click on the template icon to open a dropdown menu:
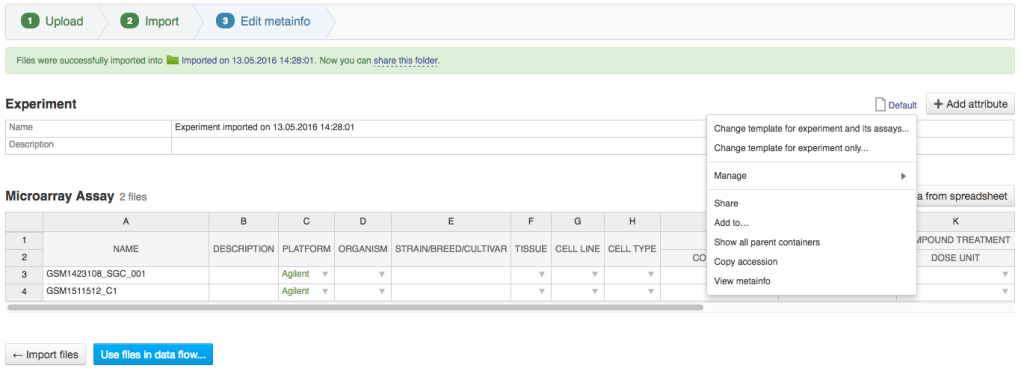
2) anytime after the files have been uploaded: right-click on your file and select Edit metainfo app:
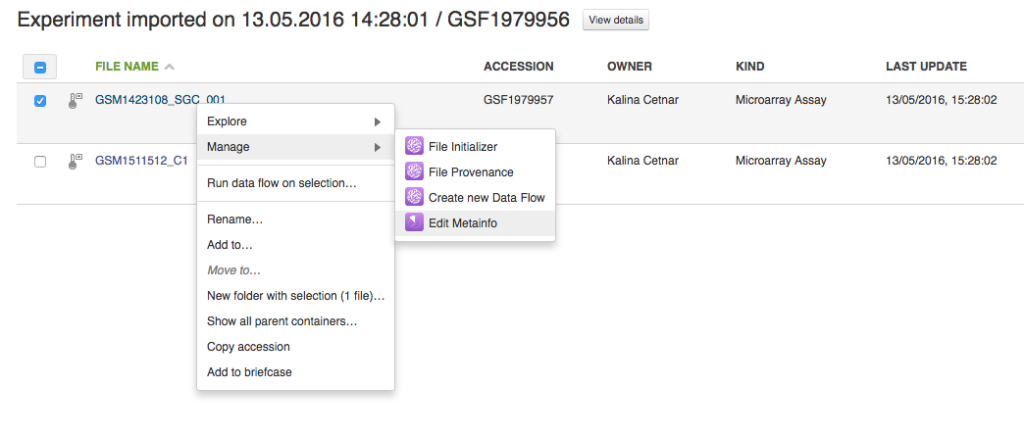
It will take you to the Edit metainfo app page. Clicking on the template icon will open a dropdown menu where you can select "Change template":

Read more about importing data into Genestack
here.
Whole Exome Sequencing Public Data Flow
Another change we made to the platform is addition of a new public data flow:
Whole Exome Sequencing Data Flow.
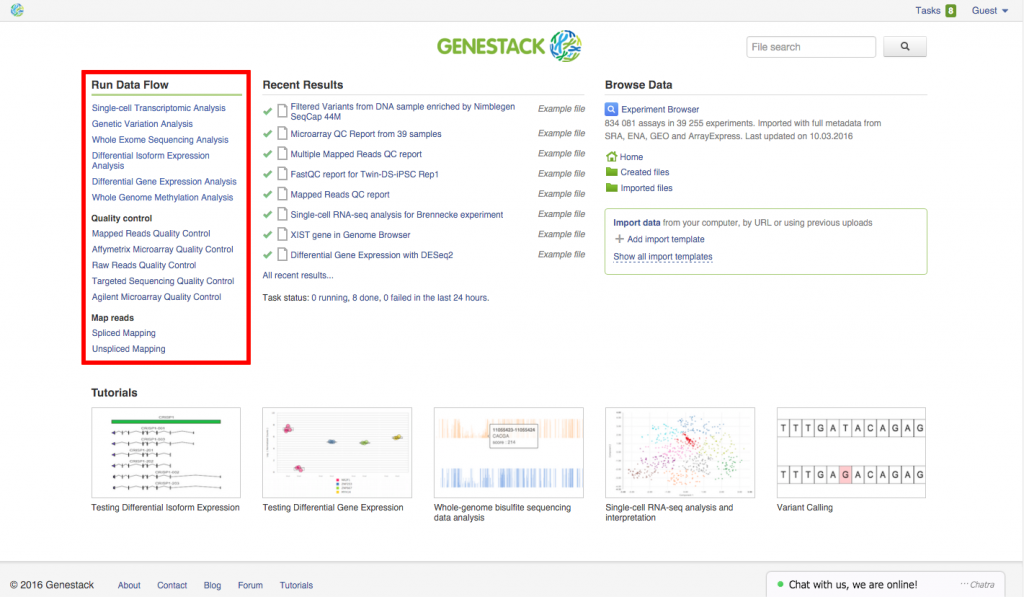
This data flow can be accessed straight from the Welcome Page, along with other useful public data flows (e.g.
Raw Reads Quality Control public data flow). You can read more about the particular steps included in this data flow in out
Whole Exome Sequencing Analysis Tutorial. To read more about data flows and how to use and create them, go to our
Getting Started Tutorial. These are the most important changes we have introduced since the
last Platform Release note. You might notice with the current release note, we've introduced a new numbering scheme. Keep tabs on
Genestack and stay tuned for more exciting features and updates! If you have any questions or suggestions, please feel free to leave them in comments below or
email us. Genestack team

 2) anytime after the files have been uploaded: right-click on your file and select Edit metainfo app:
2) anytime after the files have been uploaded: right-click on your file and select Edit metainfo app:  It will take you to the Edit metainfo app page. Clicking on the template icon will open a dropdown menu where you can select "Change template":
It will take you to the Edit metainfo app page. Clicking on the template icon will open a dropdown menu where you can select "Change template":  Read more about importing data into Genestack here.
Read more about importing data into Genestack here.  This data flow can be accessed straight from the Welcome Page, along with other useful public data flows (e.g. Raw Reads Quality Control public data flow). You can read more about the particular steps included in this data flow in out Whole Exome Sequencing Analysis Tutorial. To read more about data flows and how to use and create them, go to our Getting Started Tutorial. These are the most important changes we have introduced since the last Platform Release note. You might notice with the current release note, we've introduced a new numbering scheme. Keep tabs on Genestack and stay tuned for more exciting features and updates! If you have any questions or suggestions, please feel free to leave them in comments below or email us. Genestack team
This data flow can be accessed straight from the Welcome Page, along with other useful public data flows (e.g. Raw Reads Quality Control public data flow). You can read more about the particular steps included in this data flow in out Whole Exome Sequencing Analysis Tutorial. To read more about data flows and how to use and create them, go to our Getting Started Tutorial. These are the most important changes we have introduced since the last Platform Release note. You might notice with the current release note, we've introduced a new numbering scheme. Keep tabs on Genestack and stay tuned for more exciting features and updates! If you have any questions or suggestions, please feel free to leave them in comments below or email us. Genestack team