Building a Bioinformatics Assistant
Introduction
In this brief article, the first of a series of three brief articles, we share some of our direct experience using Large Language Models (LLMs) in life science R&D. Within each article we suggest three key practical, applicable takeaways to consider when you start using this technology.
There are many ways to describe and understand LLMs. For the purposes of this discussion we will view LLMs as an enabling technology that can be used in software projects in life sciences. Thus, rather than diving deep into what LLMs are, we should consider what they are capable of doing.
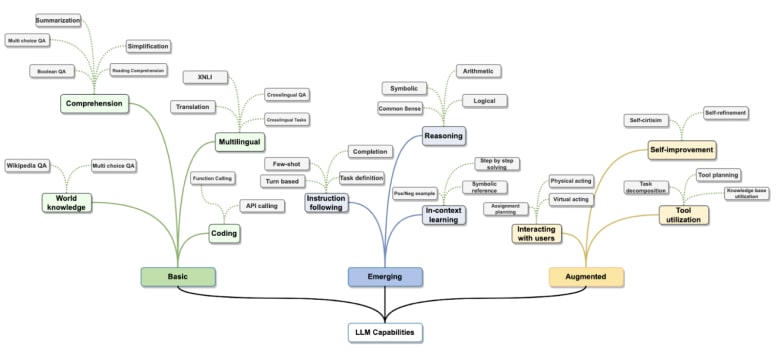
If you would like to know more about LLMs intrinsically, there are a number of excellent resources we can recommend. For a non-technical introduction to language models, we suggest this 60 minute video by Andrej Karpathy, one of the foremost computer scientists in AI, a co-founder of OpenAI. A (much) more technical introduction is available in this Feb 2024 Large Language Models: A Survey paper. The figure below, from that paper, provides a high-level overview of LLM capabilities.
A broad overview of LLM capabilities (Figure from Minaee et al, 2004.) Click to zoom
However, the key, fundamental capability we, as users of the technology, ought to understand is that it is a technology that enables computers to understand natural language. In these notes, we will look at attempts to use this technology in three specific ways as applied to life science R&D.
- Tool Utilisation: Building an LLM-Powered Bioinformatics Assistant (this note)
- Data Analysis: Using LLMs with Large Scale Biodata
- Knowledge Exploration: Working with Public & Proprietary Databases
Tool Utilisation: Building a Bioinformatics Assistant
LLMs and External Tools, a Primer
We will discuss in this note the idea of building an assistant tool for bioinformatics data processing using an LLM as an engine to translate user queries into uses of computation tools. First we provide a bit of background to familiarise you with the ideas involved. As explained, LLMs understand natural language. When provided with a prompt such as “What is the function of IL6?” an LLM will respond with text. For instance:
ChatGPT 4 responds to a simple prompt (retrieved 14 March 2024). Click to zoom
In this example ChatGPT 4 provided an answer, based on the general world knowledge that it was trained on, including Wikipedia articles, various web pages, and other resources. However, what if the user asked about something that the model doesn’t know about, such as, say, the time “right now”, or the current stock prices, or the very latest knowledge about IL-6? This is where tools come in, and the way this works is by modifying the user’s query with additional information. We add to the user’s query additional text, instructing the LLM that when it’s generating a response to the query, it can, instead of generating an answer to the user’s question, produce a request to call an external tool, if there’s a tool that appears suitable out of a list of tools, which we also provide.
Thus, a simple query “What is the function of IL-6?” might become something like this:
Note, that the user would still only enter in the original query, in boldface above, the additional text with the tools and instructions is added without exposing that to the user. Now, if we send the above query to the model, we receive quite a different response:
Now, the model outputs just one line, effectively telling the user to call the “Gene Function Lookup Tool” with one input argument, “IL-6”. All that remains is for us, before showing the user the model’s output, to parse it and, if there’s an instruction to call a tool, to do so, and show the tool’s output. The tool in question could simply be the lookup of a gene’s function in a public database, say, in Ensembl. We will discuss using LLMs with databases in a later article, but here we will consider using LLMs to call complex computational tools, such as commonly used bioinformatics algorithms, in Python or R, for data processing.
Three Tips for Using LLMs with External Tools for Bioinformatics Data Analysis
So, you’re excited by the promise of a computer that understands natural language and can call various tools. An obvious use case is to use this capability to enable non-technical users, e.g., in life sciences, this might be a biologist, to do data analysis. You envision the following scenario:
- User uploads a data spreadsheet
- User specifies what kind of analysis is required
- LLM identifies the most suitable tool and calls it
- User is presented with tool output
- LLM produces a natural language interpretation of tool output
This scenario is quite similar to what you might find in leading LLM vendor interfaces, including OpenAI’s ChatGPT 4. It is possible to implement something like this for bioinformatics R&D. However, based on our experience, we’d like to share the following recommendations/findings. Note, that we will assume in this text that you have figured out how to deliver the user’s data to your tool-augmented LLM. There are separate considerations about this point, and we will deal with them in the second article in this series (“Data Analysis: Using LLMs with Large Scale BioData”).
Tip 1. You have to make a choice: power & flexibility versus reliability & transparency
There are, broadly speaking, two ways to augment an LLM with analytical capabilities: use it to generate and execute code, or use it to launch one or more from a list of predefined tools. Below is a brief explanation and a comparison of these two approaches.
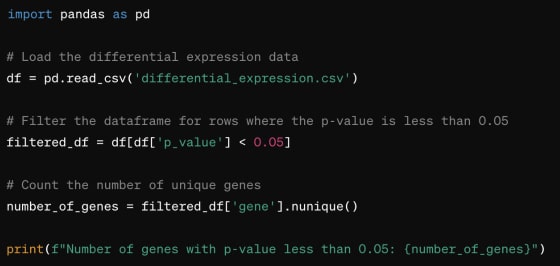
The idea is straightforward. The user asks something like “How many genes in this differential expression analysis table have p-value < 0.05?”
Code Generation
If we are using the code generation approach, we might provide the LLM with the following tool description:
Tool name: Python Runner
Tool argument: Python Code to Run
And we might also put into the LLM prompt the location and description of the data that the user has uploaded.
If an LLM was trained to generate Python (and most modern big commercially available ones are), it would, in response to a user’s query, generate a chunk of Python code, something like this:
We would then run this code, and either display the output directly to the user, or feed the tool’s output back to the LLM and ask it to rephrase or interpret it. Or both.
Predefined Tools
If we are using the predefined tool approach, such a tool might be described as follows:
Tool name: Gene Filter
Tool argument: pValueCutoff
And the model might simply request that we call the Gene Filter tool with the single argument 0.05. (You would need to figure out how to provide the data to this tool, of course.) Below is a comparison of the two approaches. One way or another, you’ll need to decide which one to adopt.
| Code Generation | Predefined Tools | |
|---|---|---|
| Advantages | Can generate code to perform just about any task the user requests, hugely expanding the capability of the LLM assistant. Does not require any curation or selection of tools to use in the assistant, can get to “working prototype” significantly faster. Your LLM system prompt will be quite short and clear. | Can perform a limited number of tasks, but always highly reliably, with guaranteed outcomes and interpretability. |
| Downsides and limitations | Libraries LLMs will generate code which will require libraries which might not be preinstalled on your system. This is unavoidable. You will thus need to either (1) detect the use of these libraries and install them on the fly, or (2) preinstall libraries beforehand and constrain the LLM by instructing it only to use libraries from a given list. The former option adds significant complexity to your implementation (a library might fail to install, might be incompatible with your version of R or Python, or might be unavailable for your system, or be unsafe). That said, in our experience, two-three dozen common libraries seems to suffice for most generated code. The latter is a more practical approach, but it too adds to the complexity. Your LLM system prompt will get more and more complex. Correctness & Interpretability There is no mechanism for you to check that the code generated is correct, and if you’re planning to build a bioinformatics assistant which allows users to run arbitrary analyses, the LLM will generate increasingly complex code. E.g., it might use incorrect mathematical formulas, make algorithmic mistakes, and so forth. You can mitigate by displaying the code generated to the user (as ChatGPT 4 does, for instance), you need to take into account that the user may not have sufficient competency to verify its correctness. Errors Arbitrary code generated by the LLM will not run correctly in all cases. In fact, in our experience, even with the more advanced models available today, it is clear that the frequency with which errors creates two issues: (1) data analysis runtimes tend to increase significantly since code must be re-generated and re-run after an error often several times until the error is resolved, impairing the user experience, and (2) the user interface must be built to accommodate the errors and occasional complete failures to complete the task. | You will need to decide what set of capabilities your assistant will have. You will need to expose these capabilities to your assistant, either by implementing each capability by hand, or by providing a set of ready-to-go APIs for these capabilities. In either case, you will need to decide what will be in this set and this decision will impose hard constraints on what your assistant will be able to do. This approach will take you longer to get to “working prototype” because you’ll need to not only implement a fairly broad collection of external tools but also test that they work with the LLM by themselves and in combinations. Testing such a system is challenging, because even with a predefined set of external tools, LLM output is non-deterministic and starting with the same scenarios you might get slightly different outcomes. This approach will consume a fair chunk of the context, because all the tool descriptions will form part of the system prompt to the LLM. You will need to deal with a complex user experience challenge. LLM capability as provided through various chat/copilot interfaces has created an expectation of infinite capability and knowledge, while this assistant will be quite severely constrained, frequently telling the user “I do not know how to do that.” |
On balance, while the potential for the “Code Generation” approach is much higher, unless you have significant resources to invest into it, and handle the complexities involved, we recommend the predefined tools approach.
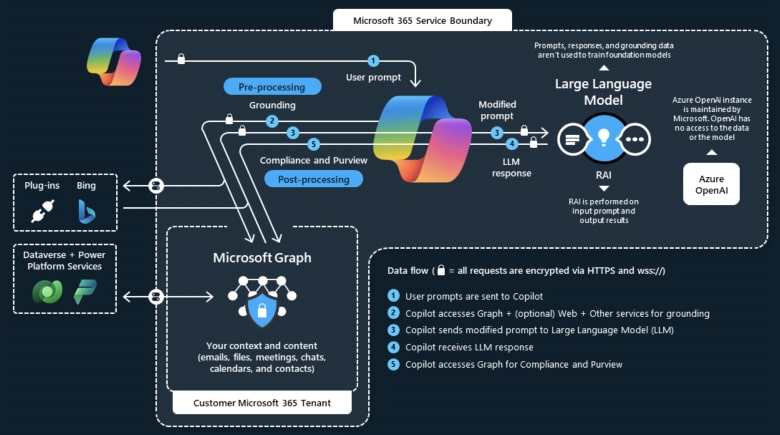
That said, there has been significant progress in recent times in creating custom language models trained to output something between general Python code and calls to external tools, in other words, to produce code in a domain specific language, which then is translated into verified, executable code. This is one of the approaches taken by Microsoft in its Copilot (see figure below). As industry develops, such approaches may become more accessible and standardised.
Tip 2: Ensure your tools have clean, self-documenting APIs for input and for output
Now that you have decided to go down the predefined tools route (right?) you will need to expose your LLM to a lot of tools. How do you do that? There are many ways to do that, from simple inclusions into the prompt as suggested above, to using specialised API’s such as function calling to providing tools as Actions directly to a pre-packaged model using the GPTs framework from OpenAI.
Regardless of how you do it, you’ll need to come up with a description of your tools to feed to the LLM. And, all of a sudden, you find yourself in the world of Data Management. Except that rather than managing datasets of assay measurements, or clinical trial records, or digital biomarkers (we will get to those), your dataset is your collection of tools. And you’re likely to end up with a lot of them – tens, and possibly hundreds.
The first surprising fact is that, in our experience, creating a collection of well-described tools of the right level of granularity for an LLM to use, with consistent natural language descriptions, is a highly non-trivial task. There is an interesting trade-off in play: on the one hand, the programming language itself, say, Python or R, together with most commonly used libraries, say, numpy, scipy and matplotlib, and some biodata-specific ones like BioConductor for R, is itself that collection of tools. They are well-documented, in terms of inputs and outputs. Why not simply expose every module and function of these tools to the LLM?
Well there are two problems with this. First: the LLM already knows about them – all that documentation and knowledge was present in the general training knowledge that went into the model, together with a lot of code that uses these libraries. That’s the reason the LLM can generate useful code to begin with. Second: there are too many there! You’ll quickly see that this approach rapidly overwhelms even the models with the largest possible context. And then there’s the third problem: these descriptions might not work well enough at all for your purposes.
Thus you’re back to curating your dataset of tools, whether you like it or not. While this article is not about the many important aspects of data curation, we will highlight a few important points:
- Create a system to track your tools. You will need a versioned tool database. It will be a challenge to track.
- Ensure you have clean, consistent descriptions of your tools. Test these with your LLM. You won’t get it right on the first try.
- Don’t forget to describe your tool outputs well!
The last point is not obvious at first glance. After all, the LLM will call your tool and to do that it needs to have the tool name and its input arguments only, right? Well, yes indeed, but one of the first things that you will discover is that once you have produced, and fed back to the LLM the output of a tool, the user will want to run further tools on that output.
“Compute summary statistics on this big table of numbers, for each column, and use these to create a box plot of maxima/minima/interquartile ranges and medians.”
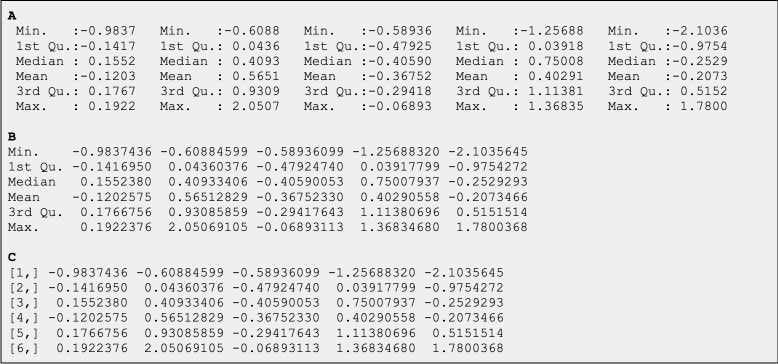
Suppose your user puts this query to your assistant, an LLM empowered with a set of predefined tools. You have a tool to compute summary statistics, and a tool to draw box plots. Your user uploads a table and… the LLM will easily identify the first tool to run. Now, consider the following ways to output a set of summary statistics for a 5 column.
All three are outputs of different calls in R. They produce the same values. But, as you might suspect, output C provides the least amount of information to the LLM, and it is likely to struggle significantly figuring out how to shape the values for the box plot. Which row contains the means and which the medians? It is important to label the outputs well. And when the outputs are so big that they don’t fit into the the LLM context and have to be stored in a file, you will need to either provide a sample of the file to the LLM context, again well-labelled, or you will need to include a clean description of the output of your tool right there with a description of it function and input arguments.
This becomes especially important if your tools, as they often do in bioinformatics, produce lists of accessions or identifiers, such as gene or protein names. They might come from incompatible sources and may need to be mapped. We will talk about making LLMs with external databases in our third article.
Tip 3: Don’t worry about the number of tools you might need
One of the things you might start to worry about now that you’ve got this far is – I have quite a lot of tools, and I’m starting to worry that (1) I’ll start running out of LLM context and (2) as the number of tools grows, will the LLM know which tools to select for a specific task? Well, here I have nothing but good news for you, with some caveats of course.
- Yes you will at some point start running into these problems! However, as of the writing of this article, context windows of modern LLMs are rapidly growing with 1-10M token contexts becoming generally available. If the description of one tool takes a hundred or so tokens, you could easily fit a hundred tools into 10,000 tokens.
- Models are getting better and better at finding the sought for text in an ever larger context. While until recently it has been observed that models tend to retrieve information better from the first and last 25% of the context window, more modern models such as Google’s Gemini 1.5 and Antrhopic’s Claude 2 and 3 are reported to be consistent at retrieving uniformly across 1-10M token window lengths (Google’s Gemini report, 2024).
- If the above two assurances are insufficient, we believe an adopted RAG (Retrieval Augmented Generation) approach over a curated database of tool descriptions will perform well for this particular challenge.
Conclusion
We reviewed in this article some of the challenges you might encounter if you set out to build an LLM powered assistant for a specific domain, in our case, bioinformatics. The challenges are non-trivial but can be overcome. One of our main observations is that the LLM technology, while widely available, nevertheless requires a set of specialised approaches and know-how. There is a requirement of deep domain awareness to build tools that users will trust and find reliable. Together with the technical expertise to deliver this there is an additional significant surprise along the way that we have only started to touch upon in our discussion of managing tools as data. You will need a firm grasp of good data management practices, ranging from data selection and curation to data versioning, governance and maintenance. (Something we can help make the easiest part of your challenges thankfully.)
Working with LLMs, like any new technology, is a learning curve. In this first of three articles we have covered some of our learnings from working with them in practical applications in Life Sciences. In the next two articles we will share our knowledge on how to work with the data itself and the more complex issue of databases.
Want to make LLMs/AI implementation simple and straightforward? Don’t want to wait for the other articles? We are here to help. You can find out more information at www.genestack.com/ai or reach out for a conversation to .